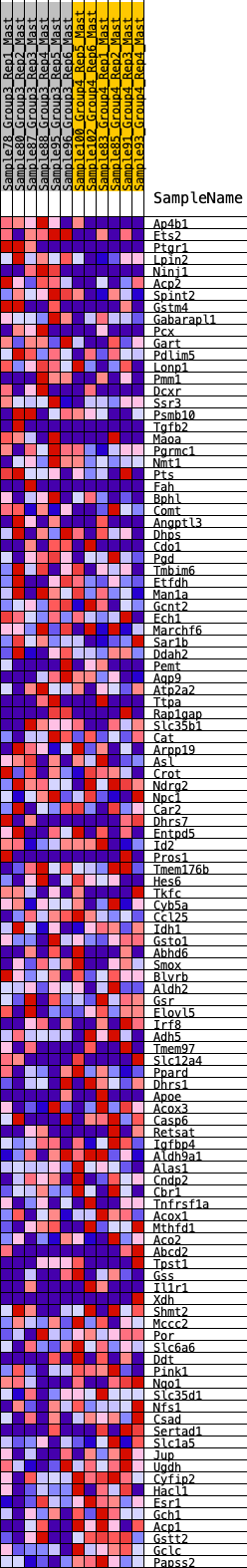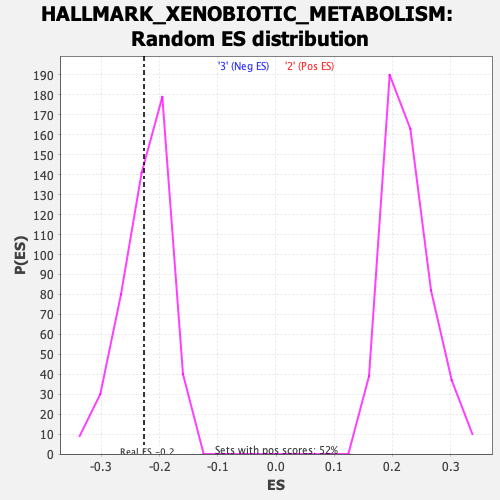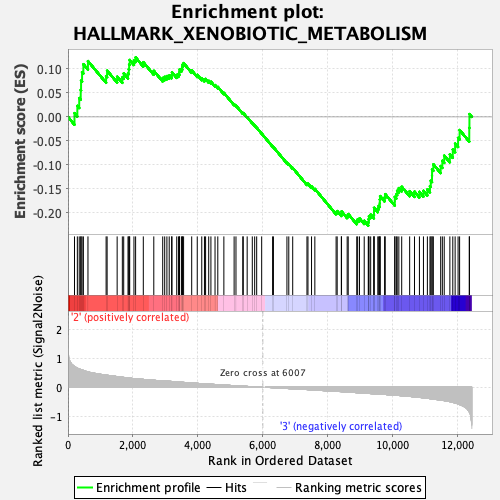
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.22667427 |
| Normalized Enrichment Score (NES) | -1.0104166 |
| Nominal p-value | 0.39874738 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ap4b1 | 200 | 0.716 | 0.0079 | No |
| 2 | Ets2 | 288 | 0.662 | 0.0230 | No |
| 3 | Ptgr1 | 346 | 0.624 | 0.0394 | No |
| 4 | Lpin2 | 392 | 0.610 | 0.0563 | No |
| 5 | Ninj1 | 403 | 0.608 | 0.0759 | No |
| 6 | Acp2 | 434 | 0.597 | 0.0935 | No |
| 7 | Spint2 | 474 | 0.580 | 0.1099 | No |
| 8 | Gstm4 | 615 | 0.532 | 0.1164 | No |
| 9 | Gabarapl1 | 1174 | 0.416 | 0.0851 | No |
| 10 | Pcx | 1205 | 0.414 | 0.0966 | No |
| 11 | Gart | 1514 | 0.366 | 0.0840 | No |
| 12 | Pdlim5 | 1674 | 0.344 | 0.0827 | No |
| 13 | Lonp1 | 1716 | 0.339 | 0.0907 | No |
| 14 | Pmm1 | 1849 | 0.320 | 0.0908 | No |
| 15 | Dcxr | 1873 | 0.318 | 0.0997 | No |
| 16 | Ssr3 | 1885 | 0.317 | 0.1094 | No |
| 17 | Psmb10 | 1901 | 0.315 | 0.1188 | No |
| 18 | Tgfb2 | 2032 | 0.298 | 0.1183 | No |
| 19 | Maoa | 2083 | 0.294 | 0.1241 | No |
| 20 | Pgrmc1 | 2320 | 0.271 | 0.1141 | No |
| 21 | Nmt1 | 2643 | 0.242 | 0.0961 | No |
| 22 | Pts | 2918 | 0.219 | 0.0813 | No |
| 23 | Fah | 2979 | 0.214 | 0.0836 | No |
| 24 | Bphl | 3047 | 0.212 | 0.0853 | No |
| 25 | Comt | 3118 | 0.207 | 0.0866 | No |
| 26 | Angptl3 | 3192 | 0.200 | 0.0874 | No |
| 27 | Dhps | 3204 | 0.199 | 0.0932 | No |
| 28 | Cdo1 | 3350 | 0.186 | 0.0877 | No |
| 29 | Pgd | 3403 | 0.183 | 0.0896 | No |
| 30 | Tmbim6 | 3432 | 0.179 | 0.0933 | No |
| 31 | Etfdh | 3437 | 0.178 | 0.0990 | No |
| 32 | Man1a | 3495 | 0.173 | 0.1002 | No |
| 33 | Gcnt2 | 3520 | 0.171 | 0.1040 | No |
| 34 | Ech1 | 3529 | 0.170 | 0.1091 | No |
| 35 | Marchf6 | 3561 | 0.168 | 0.1122 | No |
| 36 | Sar1b | 3811 | 0.148 | 0.0970 | No |
| 37 | Ddah2 | 3982 | 0.135 | 0.0878 | No |
| 38 | Pemt | 4121 | 0.125 | 0.0808 | No |
| 39 | Aqp9 | 4204 | 0.119 | 0.0781 | No |
| 40 | Atp2a2 | 4239 | 0.116 | 0.0793 | No |
| 41 | Ttpa | 4335 | 0.110 | 0.0752 | No |
| 42 | Rap1gap | 4403 | 0.106 | 0.0734 | No |
| 43 | Slc35b1 | 4530 | 0.096 | 0.0664 | No |
| 44 | Cat | 4615 | 0.090 | 0.0626 | No |
| 45 | Arpp19 | 4802 | 0.077 | 0.0501 | No |
| 46 | Asl | 5118 | 0.056 | 0.0264 | No |
| 47 | Crot | 5175 | 0.051 | 0.0236 | No |
| 48 | Ndrg2 | 5385 | 0.038 | 0.0079 | No |
| 49 | Npc1 | 5399 | 0.037 | 0.0081 | No |
| 50 | Car2 | 5520 | 0.030 | -0.0006 | No |
| 51 | Dhrs7 | 5678 | 0.020 | -0.0127 | No |
| 52 | Entpd5 | 5751 | 0.016 | -0.0180 | No |
| 53 | Id2 | 5813 | 0.012 | -0.0225 | No |
| 54 | Pros1 | 5967 | 0.002 | -0.0348 | No |
| 55 | Tmem176b | 6303 | -0.012 | -0.0616 | No |
| 56 | Hes6 | 6329 | -0.015 | -0.0631 | No |
| 57 | Tkfc | 6745 | -0.041 | -0.0954 | No |
| 58 | Cyb5a | 6802 | -0.044 | -0.0985 | No |
| 59 | Ccl25 | 6923 | -0.052 | -0.1065 | No |
| 60 | Idh1 | 7362 | -0.078 | -0.1394 | No |
| 61 | Gsto1 | 7393 | -0.080 | -0.1391 | No |
| 62 | Abhd6 | 7503 | -0.086 | -0.1451 | No |
| 63 | Smox | 7605 | -0.093 | -0.1501 | No |
| 64 | Blvrb | 8260 | -0.138 | -0.1985 | No |
| 65 | Aldh2 | 8293 | -0.140 | -0.1964 | No |
| 66 | Gsr | 8421 | -0.149 | -0.2017 | No |
| 67 | Elovl5 | 8427 | -0.149 | -0.1971 | No |
| 68 | Irf8 | 8601 | -0.163 | -0.2057 | No |
| 69 | Adh5 | 8633 | -0.166 | -0.2026 | No |
| 70 | Tmem97 | 8896 | -0.184 | -0.2176 | No |
| 71 | Slc12a4 | 8924 | -0.187 | -0.2136 | No |
| 72 | Ppard | 8980 | -0.190 | -0.2116 | No |
| 73 | Dhrs1 | 9126 | -0.201 | -0.2166 | No |
| 74 | Apoe | 9251 | -0.210 | -0.2196 | Yes |
| 75 | Acox3 | 9262 | -0.211 | -0.2133 | Yes |
| 76 | Casp6 | 9273 | -0.212 | -0.2070 | Yes |
| 77 | Retsat | 9325 | -0.216 | -0.2038 | Yes |
| 78 | Igfbp4 | 9424 | -0.223 | -0.2043 | Yes |
| 79 | Aldh9a1 | 9428 | -0.223 | -0.1970 | Yes |
| 80 | Alas1 | 9429 | -0.223 | -0.1895 | Yes |
| 81 | Cndp2 | 9545 | -0.229 | -0.1912 | Yes |
| 82 | Cbr1 | 9572 | -0.231 | -0.1855 | Yes |
| 83 | Tnfrsf1a | 9605 | -0.234 | -0.1802 | Yes |
| 84 | Acox1 | 9607 | -0.234 | -0.1724 | Yes |
| 85 | Mthfd1 | 9616 | -0.235 | -0.1651 | Yes |
| 86 | Aco2 | 9754 | -0.246 | -0.1680 | Yes |
| 87 | Abcd2 | 9773 | -0.247 | -0.1611 | Yes |
| 88 | Tpst1 | 10068 | -0.272 | -0.1758 | Yes |
| 89 | Gss | 10069 | -0.272 | -0.1667 | Yes |
| 90 | Il1r1 | 10116 | -0.276 | -0.1612 | Yes |
| 91 | Xdh | 10142 | -0.278 | -0.1539 | Yes |
| 92 | Shmt2 | 10193 | -0.282 | -0.1484 | Yes |
| 93 | Mccc2 | 10281 | -0.292 | -0.1457 | Yes |
| 94 | Por | 10526 | -0.313 | -0.1549 | Yes |
| 95 | Slc6a6 | 10675 | -0.331 | -0.1558 | Yes |
| 96 | Ddt | 10827 | -0.349 | -0.1563 | Yes |
| 97 | Pink1 | 10951 | -0.362 | -0.1541 | Yes |
| 98 | Nqo1 | 11072 | -0.379 | -0.1511 | Yes |
| 99 | Slc35d1 | 11152 | -0.390 | -0.1444 | Yes |
| 100 | Nfs1 | 11177 | -0.394 | -0.1331 | Yes |
| 101 | Csad | 11215 | -0.398 | -0.1227 | Yes |
| 102 | Sertad1 | 11217 | -0.398 | -0.1094 | Yes |
| 103 | Slc1a5 | 11256 | -0.405 | -0.0989 | Yes |
| 104 | Jup | 11482 | -0.437 | -0.1024 | Yes |
| 105 | Ugdh | 11533 | -0.447 | -0.0915 | Yes |
| 106 | Cyfip2 | 11591 | -0.459 | -0.0806 | Yes |
| 107 | Hacl1 | 11765 | -0.494 | -0.0781 | Yes |
| 108 | Esr1 | 11855 | -0.513 | -0.0680 | Yes |
| 109 | Gch1 | 11927 | -0.543 | -0.0555 | Yes |
| 110 | Acp1 | 12018 | -0.572 | -0.0436 | Yes |
| 111 | Gstt2 | 12065 | -0.594 | -0.0274 | Yes |
| 112 | Gclc | 12363 | -0.852 | -0.0228 | Yes |
| 113 | Papss2 | 12368 | -0.863 | 0.0059 | Yes |