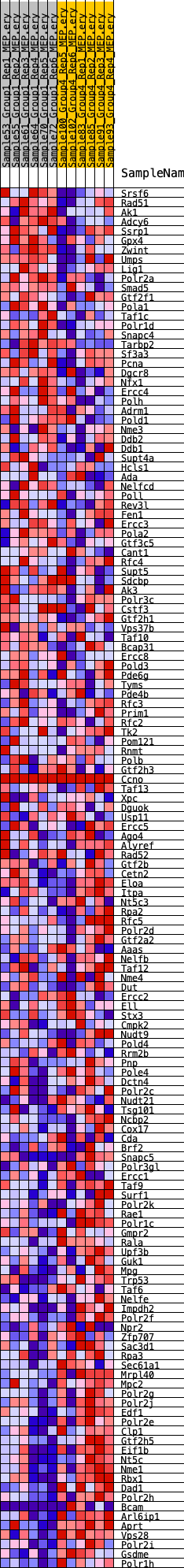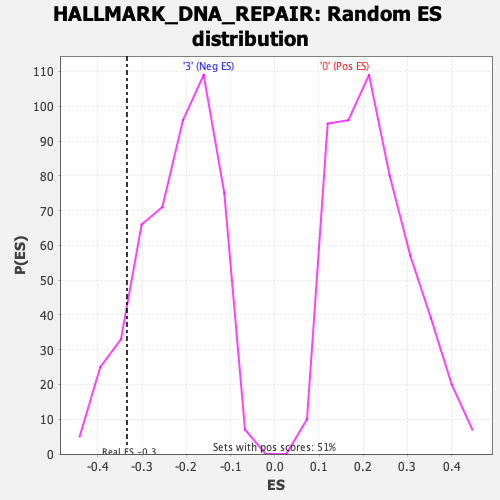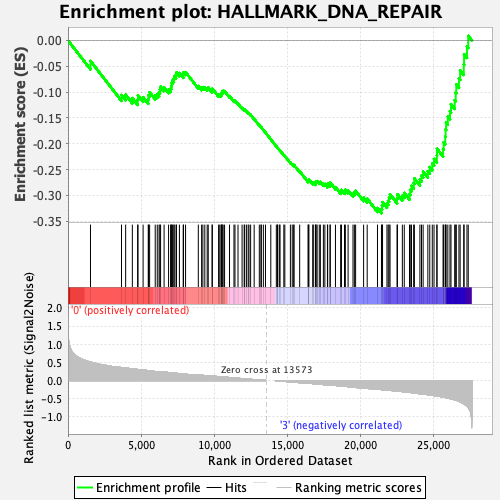
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.33448574 |
| Normalized Enrichment Score (NES) | -1.5283923 |
| Nominal p-value | 0.11088296 |
| FDR q-value | 0.45587426 |
| FWER p-Value | 0.444 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Srsf6 | 1537 | 0.507 | -0.0398 | No |
| 2 | Rad51 | 3657 | 0.356 | -0.1057 | No |
| 3 | Ak1 | 3941 | 0.338 | -0.1052 | No |
| 4 | Adcy6 | 4398 | 0.318 | -0.1117 | No |
| 5 | Ssrp1 | 4761 | 0.304 | -0.1152 | No |
| 6 | Gpx4 | 4784 | 0.303 | -0.1063 | No |
| 7 | Zwint | 5133 | 0.286 | -0.1099 | No |
| 8 | Umps | 5485 | 0.272 | -0.1140 | No |
| 9 | Lig1 | 5504 | 0.271 | -0.1061 | No |
| 10 | Polr2a | 5579 | 0.266 | -0.1003 | No |
| 11 | Smad5 | 5962 | 0.248 | -0.1063 | No |
| 12 | Gtf2f1 | 6102 | 0.242 | -0.1036 | No |
| 13 | Pola1 | 6224 | 0.237 | -0.1005 | No |
| 14 | Taf1c | 6284 | 0.234 | -0.0952 | No |
| 15 | Polr1d | 6329 | 0.232 | -0.0894 | No |
| 16 | Snapc4 | 6568 | 0.228 | -0.0908 | No |
| 17 | Tarbp2 | 6874 | 0.220 | -0.0949 | No |
| 18 | Sf3a3 | 7012 | 0.214 | -0.0931 | No |
| 19 | Pcna | 7059 | 0.211 | -0.0881 | No |
| 20 | Dgcr8 | 7090 | 0.210 | -0.0825 | No |
| 21 | Nfx1 | 7137 | 0.208 | -0.0775 | No |
| 22 | Ercc4 | 7232 | 0.204 | -0.0745 | No |
| 23 | Polh | 7261 | 0.204 | -0.0690 | No |
| 24 | Adrm1 | 7383 | 0.199 | -0.0671 | No |
| 25 | Pold1 | 7415 | 0.198 | -0.0619 | No |
| 26 | Nme3 | 7615 | 0.190 | -0.0631 | No |
| 27 | Ddb2 | 7875 | 0.179 | -0.0669 | No |
| 28 | Ddb1 | 7893 | 0.179 | -0.0618 | No |
| 29 | Supt4a | 8036 | 0.174 | -0.0614 | No |
| 30 | Hcls1 | 8907 | 0.152 | -0.0883 | No |
| 31 | Ada | 9132 | 0.144 | -0.0918 | No |
| 32 | Nelfcd | 9219 | 0.142 | -0.0905 | No |
| 33 | Poll | 9351 | 0.137 | -0.0909 | No |
| 34 | Rev3l | 9514 | 0.132 | -0.0926 | No |
| 35 | Fen1 | 9586 | 0.129 | -0.0910 | No |
| 36 | Ercc3 | 9851 | 0.122 | -0.0968 | No |
| 37 | Pola2 | 9862 | 0.121 | -0.0933 | No |
| 38 | Gtf3c5 | 10293 | 0.107 | -0.1055 | No |
| 39 | Cant1 | 10374 | 0.105 | -0.1051 | No |
| 40 | Rfc4 | 10462 | 0.101 | -0.1050 | No |
| 41 | Supt5 | 10489 | 0.100 | -0.1028 | No |
| 42 | Sdcbp | 10529 | 0.099 | -0.1011 | No |
| 43 | Ak3 | 10541 | 0.099 | -0.0983 | No |
| 44 | Polr3c | 10605 | 0.096 | -0.0976 | No |
| 45 | Cstf3 | 10702 | 0.093 | -0.0981 | No |
| 46 | Gtf2h1 | 11041 | 0.082 | -0.1078 | No |
| 47 | Vps37b | 11344 | 0.071 | -0.1165 | No |
| 48 | Taf10 | 11413 | 0.068 | -0.1168 | No |
| 49 | Bcap31 | 11621 | 0.062 | -0.1224 | No |
| 50 | Ercc8 | 11900 | 0.053 | -0.1308 | No |
| 51 | Pold3 | 12043 | 0.048 | -0.1345 | No |
| 52 | Pde6g | 12074 | 0.047 | -0.1341 | No |
| 53 | Tyms | 12194 | 0.043 | -0.1371 | No |
| 54 | Pde4b | 12314 | 0.039 | -0.1402 | No |
| 55 | Rfc3 | 12370 | 0.037 | -0.1410 | No |
| 56 | Prim1 | 12483 | 0.033 | -0.1440 | No |
| 57 | Rfc2 | 12725 | 0.025 | -0.1520 | No |
| 58 | Tk2 | 13077 | 0.016 | -0.1642 | No |
| 59 | Pom121 | 13188 | 0.012 | -0.1679 | No |
| 60 | Rnmt | 13216 | 0.011 | -0.1685 | No |
| 61 | Polb | 13354 | 0.008 | -0.1732 | No |
| 62 | Gtf2h3 | 13498 | 0.003 | -0.1784 | No |
| 63 | Ccno | 13862 | 0.000 | -0.1916 | No |
| 64 | Taf13 | 14240 | -0.003 | -0.2052 | No |
| 65 | Xpc | 14318 | -0.005 | -0.2079 | No |
| 66 | Dguok | 14334 | -0.006 | -0.2083 | No |
| 67 | Usp11 | 14446 | -0.009 | -0.2120 | No |
| 68 | Ercc5 | 14494 | -0.011 | -0.2134 | No |
| 69 | Ago4 | 14753 | -0.020 | -0.2221 | No |
| 70 | Alyref | 14821 | -0.023 | -0.2238 | No |
| 71 | Rad52 | 15201 | -0.036 | -0.2365 | No |
| 72 | Gtf2b | 15327 | -0.041 | -0.2397 | No |
| 73 | Cetn2 | 15427 | -0.045 | -0.2419 | No |
| 74 | Eloa | 15451 | -0.046 | -0.2413 | No |
| 75 | Itpa | 15836 | -0.057 | -0.2534 | No |
| 76 | Nt5c3 | 16414 | -0.073 | -0.2721 | No |
| 77 | Rpa2 | 16428 | -0.073 | -0.2703 | No |
| 78 | Rfc5 | 16469 | -0.075 | -0.2694 | No |
| 79 | Polr2d | 16729 | -0.084 | -0.2761 | No |
| 80 | Gtf2a2 | 16766 | -0.085 | -0.2747 | No |
| 81 | Aaas | 16907 | -0.090 | -0.2770 | No |
| 82 | Nelfb | 16927 | -0.091 | -0.2748 | No |
| 83 | Taf12 | 16958 | -0.092 | -0.2730 | No |
| 84 | Nme4 | 17028 | -0.094 | -0.2725 | No |
| 85 | Dut | 17175 | -0.100 | -0.2746 | No |
| 86 | Ercc2 | 17251 | -0.103 | -0.2741 | No |
| 87 | Ell | 17456 | -0.110 | -0.2780 | No |
| 88 | Stx3 | 17557 | -0.114 | -0.2780 | No |
| 89 | Cmpk2 | 17734 | -0.121 | -0.2805 | No |
| 90 | Nudt9 | 17750 | -0.122 | -0.2772 | No |
| 91 | Pold4 | 17915 | -0.127 | -0.2791 | No |
| 92 | Rrm2b | 17921 | -0.127 | -0.2753 | No |
| 93 | Pnp | 18282 | -0.135 | -0.2841 | No |
| 94 | Pole4 | 18637 | -0.148 | -0.2923 | No |
| 95 | Dctn4 | 18692 | -0.150 | -0.2895 | No |
| 96 | Polr2c | 18900 | -0.158 | -0.2920 | No |
| 97 | Nudt21 | 18958 | -0.160 | -0.2890 | No |
| 98 | Tsg101 | 19145 | -0.168 | -0.2904 | No |
| 99 | Ncbp2 | 19481 | -0.182 | -0.2968 | No |
| 100 | Cox17 | 19575 | -0.186 | -0.2943 | No |
| 101 | Cda | 19660 | -0.189 | -0.2913 | No |
| 102 | Brf2 | 20206 | -0.207 | -0.3046 | No |
| 103 | Snapc5 | 20451 | -0.215 | -0.3066 | No |
| 104 | Polr3gl | 21154 | -0.238 | -0.3246 | No |
| 105 | Ercc1 | 21426 | -0.248 | -0.3266 | Yes |
| 106 | Taf9 | 21465 | -0.250 | -0.3200 | Yes |
| 107 | Surf1 | 21500 | -0.252 | -0.3132 | Yes |
| 108 | Polr2k | 21802 | -0.260 | -0.3159 | Yes |
| 109 | Rae1 | 21884 | -0.264 | -0.3105 | Yes |
| 110 | Polr1c | 21947 | -0.267 | -0.3042 | Yes |
| 111 | Gmpr2 | 22013 | -0.270 | -0.2980 | Yes |
| 112 | Rala | 22491 | -0.288 | -0.3062 | Yes |
| 113 | Upf3b | 22521 | -0.289 | -0.2980 | Yes |
| 114 | Guk1 | 22854 | -0.304 | -0.3005 | Yes |
| 115 | Mpg | 22985 | -0.310 | -0.2953 | Yes |
| 116 | Trp53 | 23343 | -0.326 | -0.2980 | Yes |
| 117 | Taf6 | 23392 | -0.328 | -0.2893 | Yes |
| 118 | Nelfe | 23479 | -0.333 | -0.2818 | Yes |
| 119 | Impdh2 | 23627 | -0.340 | -0.2763 | Yes |
| 120 | Polr2f | 23676 | -0.342 | -0.2672 | Yes |
| 121 | Npr2 | 24054 | -0.362 | -0.2694 | Yes |
| 122 | Zfp707 | 24163 | -0.367 | -0.2616 | Yes |
| 123 | Sac3d1 | 24275 | -0.372 | -0.2538 | Yes |
| 124 | Rpa3 | 24597 | -0.389 | -0.2532 | Yes |
| 125 | Sec61a1 | 24728 | -0.398 | -0.2452 | Yes |
| 126 | Mrpl40 | 24900 | -0.409 | -0.2384 | Yes |
| 127 | Mpc2 | 25019 | -0.413 | -0.2296 | Yes |
| 128 | Polr2g | 25213 | -0.425 | -0.2230 | Yes |
| 129 | Polr2j | 25223 | -0.426 | -0.2098 | Yes |
| 130 | Edf1 | 25637 | -0.453 | -0.2104 | Yes |
| 131 | Polr2e | 25676 | -0.456 | -0.1973 | Yes |
| 132 | Clp1 | 25784 | -0.467 | -0.1863 | Yes |
| 133 | Gtf2h5 | 25810 | -0.468 | -0.1723 | Yes |
| 134 | Eif1b | 25844 | -0.471 | -0.1585 | Yes |
| 135 | Nt5c | 25963 | -0.484 | -0.1475 | Yes |
| 136 | Nme1 | 26109 | -0.496 | -0.1369 | Yes |
| 137 | Rbx1 | 26179 | -0.504 | -0.1234 | Yes |
| 138 | Dad1 | 26432 | -0.529 | -0.1157 | Yes |
| 139 | Polr2h | 26500 | -0.538 | -0.1010 | Yes |
| 140 | Bcam | 26542 | -0.543 | -0.0852 | Yes |
| 141 | Arl6ip1 | 26722 | -0.571 | -0.0736 | Yes |
| 142 | Aprt | 26804 | -0.586 | -0.0579 | Yes |
| 143 | Vps28 | 27051 | -0.640 | -0.0464 | Yes |
| 144 | Polr2i | 27074 | -0.645 | -0.0267 | Yes |
| 145 | Gsdme | 27271 | -0.701 | -0.0115 | Yes |
| 146 | Polr1h | 27358 | -0.740 | 0.0089 | Yes |