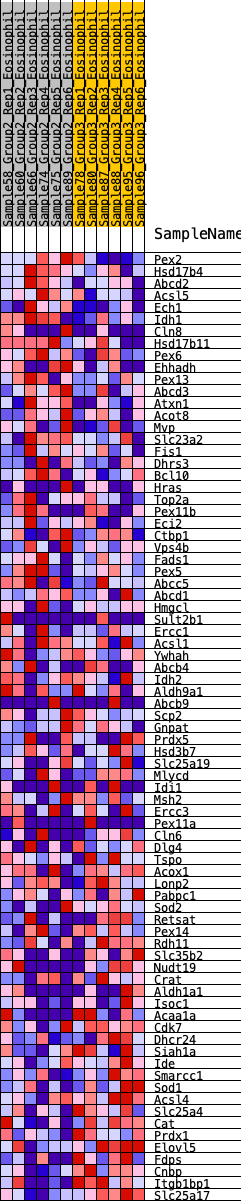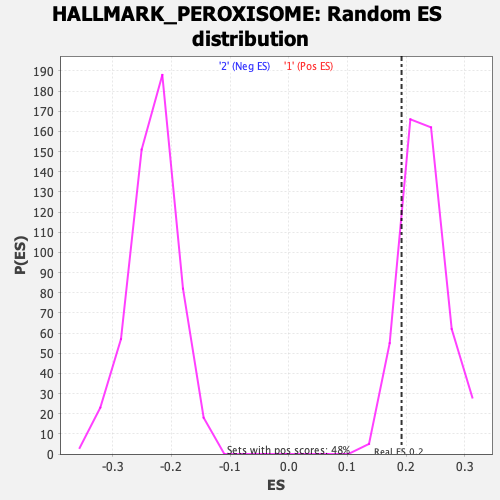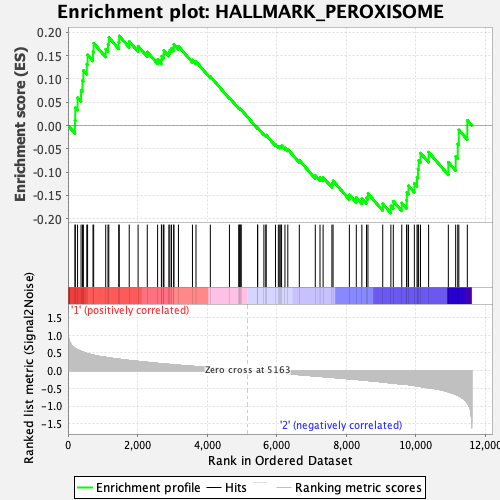
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.19217236 |
| Normalized Enrichment Score (NES) | 0.83267003 |
| Nominal p-value | 0.8619247 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex2 | 200 | 0.623 | 0.0111 | Yes |
| 2 | Hsd17b4 | 210 | 0.617 | 0.0385 | Yes |
| 3 | Abcd2 | 275 | 0.586 | 0.0597 | Yes |
| 4 | Acsl5 | 375 | 0.543 | 0.0758 | Yes |
| 5 | Ech1 | 416 | 0.529 | 0.0965 | Yes |
| 6 | Idh1 | 442 | 0.519 | 0.1180 | Yes |
| 7 | Cln8 | 541 | 0.484 | 0.1316 | Yes |
| 8 | Hsd17b11 | 563 | 0.479 | 0.1517 | Yes |
| 9 | Pex6 | 719 | 0.438 | 0.1582 | Yes |
| 10 | Ehhadh | 738 | 0.436 | 0.1766 | Yes |
| 11 | Pex13 | 1084 | 0.372 | 0.1636 | Yes |
| 12 | Abcd3 | 1150 | 0.362 | 0.1745 | Yes |
| 13 | Atxn1 | 1173 | 0.357 | 0.1889 | Yes |
| 14 | Acot8 | 1459 | 0.320 | 0.1788 | Yes |
| 15 | Mvp | 1473 | 0.318 | 0.1922 | Yes |
| 16 | Slc23a2 | 1760 | 0.284 | 0.1803 | No |
| 17 | Fis1 | 2015 | 0.259 | 0.1701 | No |
| 18 | Dhrs3 | 2279 | 0.232 | 0.1579 | No |
| 19 | Bcl10 | 2576 | 0.202 | 0.1414 | No |
| 20 | Hras | 2685 | 0.193 | 0.1409 | No |
| 21 | Top2a | 2696 | 0.193 | 0.1488 | No |
| 22 | Pex11b | 2752 | 0.188 | 0.1526 | No |
| 23 | Eci2 | 2754 | 0.188 | 0.1611 | No |
| 24 | Ctbp1 | 2900 | 0.174 | 0.1564 | No |
| 25 | Vps4b | 2929 | 0.172 | 0.1618 | No |
| 26 | Fads1 | 2976 | 0.167 | 0.1655 | No |
| 27 | Pex5 | 3040 | 0.161 | 0.1674 | No |
| 28 | Abcc5 | 3043 | 0.161 | 0.1746 | No |
| 29 | Abcd1 | 3175 | 0.151 | 0.1701 | No |
| 30 | Hmgcl | 3578 | 0.120 | 0.1407 | No |
| 31 | Sult2b1 | 3680 | 0.113 | 0.1371 | No |
| 32 | Ercc1 | 4092 | 0.081 | 0.1052 | No |
| 33 | Acsl1 | 4641 | 0.039 | 0.0594 | No |
| 34 | Ywhah | 4909 | 0.018 | 0.0371 | No |
| 35 | Abcb4 | 4924 | 0.017 | 0.0366 | No |
| 36 | Idh2 | 4944 | 0.015 | 0.0357 | No |
| 37 | Aldh9a1 | 4969 | 0.013 | 0.0342 | No |
| 38 | Abcb9 | 4978 | 0.013 | 0.0341 | No |
| 39 | Scp2 | 5450 | -0.022 | -0.0058 | No |
| 40 | Gnpat | 5453 | -0.022 | -0.0050 | No |
| 41 | Prdx5 | 5632 | -0.036 | -0.0188 | No |
| 42 | Hsd3b7 | 5690 | -0.040 | -0.0219 | No |
| 43 | Slc25a19 | 5694 | -0.041 | -0.0203 | No |
| 44 | Mlycd | 5965 | -0.060 | -0.0410 | No |
| 45 | Idi1 | 6047 | -0.066 | -0.0450 | No |
| 46 | Msh2 | 6087 | -0.068 | -0.0453 | No |
| 47 | Ercc3 | 6118 | -0.071 | -0.0446 | No |
| 48 | Pex11a | 6140 | -0.072 | -0.0431 | No |
| 49 | Cln6 | 6237 | -0.079 | -0.0478 | No |
| 50 | Dlg4 | 6319 | -0.087 | -0.0509 | No |
| 51 | Tspo | 6650 | -0.114 | -0.0743 | No |
| 52 | Acox1 | 7109 | -0.152 | -0.1071 | No |
| 53 | Lonp2 | 7245 | -0.162 | -0.1114 | No |
| 54 | Pabpc1 | 7334 | -0.169 | -0.1113 | No |
| 55 | Sod2 | 7587 | -0.190 | -0.1245 | No |
| 56 | Retsat | 7620 | -0.193 | -0.1184 | No |
| 57 | Pex14 | 8089 | -0.233 | -0.1484 | No |
| 58 | Rdh11 | 8288 | -0.248 | -0.1543 | No |
| 59 | Slc35b2 | 8448 | -0.264 | -0.1560 | No |
| 60 | Nudt19 | 8581 | -0.276 | -0.1548 | No |
| 61 | Crat | 8623 | -0.279 | -0.1457 | No |
| 62 | Aldh1a1 | 9048 | -0.322 | -0.1678 | No |
| 63 | Isoc1 | 9282 | -0.345 | -0.1722 | No |
| 64 | Acaa1a | 9352 | -0.350 | -0.1622 | No |
| 65 | Cdk7 | 9595 | -0.377 | -0.1660 | No |
| 66 | Dhcr24 | 9735 | -0.389 | -0.1603 | No |
| 67 | Siah1a | 9742 | -0.389 | -0.1431 | No |
| 68 | Ide | 9790 | -0.396 | -0.1291 | No |
| 69 | Smarcc1 | 9957 | -0.421 | -0.1242 | No |
| 70 | Sod1 | 10034 | -0.432 | -0.1111 | No |
| 71 | Acsl4 | 10061 | -0.437 | -0.0934 | No |
| 72 | Slc25a4 | 10080 | -0.440 | -0.0749 | No |
| 73 | Cat | 10134 | -0.448 | -0.0590 | No |
| 74 | Prdx1 | 10367 | -0.485 | -0.0570 | No |
| 75 | Elovl5 | 10934 | -0.600 | -0.0787 | No |
| 76 | Fdps | 11145 | -0.672 | -0.0662 | No |
| 77 | Cnbp | 11205 | -0.700 | -0.0394 | No |
| 78 | Itgb1bp1 | 11236 | -0.716 | -0.0093 | No |
| 79 | Slc25a17 | 11480 | -0.911 | 0.0112 | No |