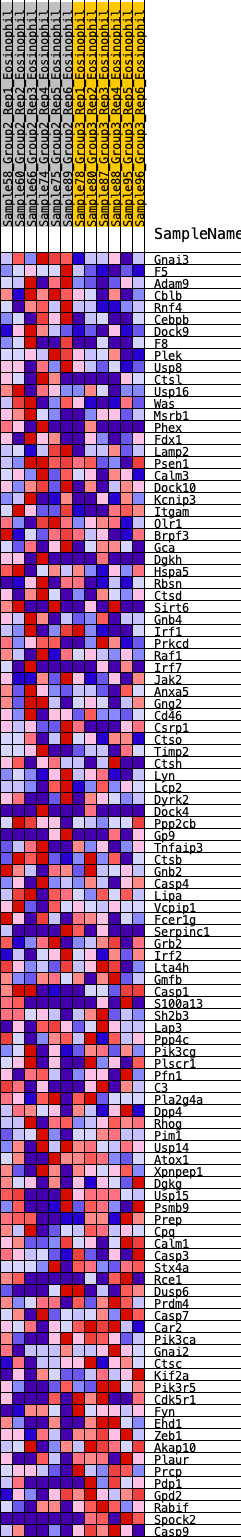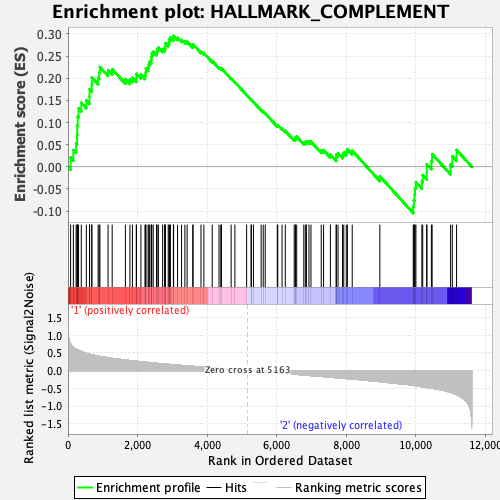
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group3.Eosinophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.29575878 |
| Normalized Enrichment Score (NES) | 1.3273822 |
| Nominal p-value | 0.038383838 |
| FDR q-value | 0.6144708 |
| FWER p-Value | 0.742 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gnai3 | 75 | 0.760 | 0.0209 | Yes |
| 2 | F5 | 154 | 0.663 | 0.0381 | Yes |
| 3 | Adam9 | 233 | 0.604 | 0.0532 | Yes |
| 4 | Cblb | 262 | 0.592 | 0.0722 | Yes |
| 5 | Rnf4 | 266 | 0.590 | 0.0932 | Yes |
| 6 | Cebpb | 280 | 0.583 | 0.1132 | Yes |
| 7 | Dock9 | 303 | 0.574 | 0.1321 | Yes |
| 8 | F8 | 382 | 0.540 | 0.1448 | Yes |
| 9 | Plek | 527 | 0.489 | 0.1500 | Yes |
| 10 | Usp8 | 618 | 0.461 | 0.1588 | Yes |
| 11 | Ctsl | 620 | 0.460 | 0.1753 | Yes |
| 12 | Usp16 | 682 | 0.447 | 0.1862 | Yes |
| 13 | Was | 686 | 0.445 | 0.2020 | Yes |
| 14 | Msrb1 | 866 | 0.406 | 0.2011 | Yes |
| 15 | Phex | 897 | 0.400 | 0.2130 | Yes |
| 16 | Fdx1 | 917 | 0.395 | 0.2256 | Yes |
| 17 | Lamp2 | 1152 | 0.361 | 0.2183 | Yes |
| 18 | Psen1 | 1270 | 0.344 | 0.2206 | Yes |
| 19 | Calm3 | 1650 | 0.296 | 0.1983 | Yes |
| 20 | Dock10 | 1780 | 0.283 | 0.1973 | Yes |
| 21 | Kcnip3 | 1852 | 0.275 | 0.2011 | Yes |
| 22 | Itgam | 1962 | 0.264 | 0.2012 | Yes |
| 23 | Olr1 | 1970 | 0.263 | 0.2101 | Yes |
| 24 | Brpf3 | 2096 | 0.251 | 0.2083 | Yes |
| 25 | Gca | 2213 | 0.238 | 0.2068 | Yes |
| 26 | Dgkh | 2235 | 0.236 | 0.2135 | Yes |
| 27 | Hspa5 | 2239 | 0.236 | 0.2218 | Yes |
| 28 | Rbsn | 2303 | 0.229 | 0.2246 | Yes |
| 29 | Ctsd | 2323 | 0.226 | 0.2311 | Yes |
| 30 | Sirt6 | 2348 | 0.224 | 0.2371 | Yes |
| 31 | Gnb4 | 2391 | 0.219 | 0.2414 | Yes |
| 32 | Irf1 | 2400 | 0.219 | 0.2486 | Yes |
| 33 | Prkcd | 2408 | 0.218 | 0.2559 | Yes |
| 34 | Raf1 | 2450 | 0.214 | 0.2600 | Yes |
| 35 | Irf7 | 2545 | 0.205 | 0.2592 | Yes |
| 36 | Jak2 | 2561 | 0.203 | 0.2653 | Yes |
| 37 | Anxa5 | 2599 | 0.200 | 0.2693 | Yes |
| 38 | Gng2 | 2721 | 0.190 | 0.2657 | Yes |
| 39 | Cd46 | 2777 | 0.185 | 0.2676 | Yes |
| 40 | Csrp1 | 2797 | 0.183 | 0.2726 | Yes |
| 41 | Ctso | 2798 | 0.183 | 0.2792 | Yes |
| 42 | Timp2 | 2881 | 0.175 | 0.2784 | Yes |
| 43 | Ctsh | 2907 | 0.173 | 0.2825 | Yes |
| 44 | Lyn | 2915 | 0.173 | 0.2881 | Yes |
| 45 | Lcp2 | 2944 | 0.170 | 0.2918 | Yes |
| 46 | Dyrk2 | 3030 | 0.163 | 0.2903 | Yes |
| 47 | Dock4 | 3036 | 0.162 | 0.2958 | Yes |
| 48 | Ppp2cb | 3150 | 0.153 | 0.2915 | No |
| 49 | Gp9 | 3264 | 0.144 | 0.2868 | No |
| 50 | Tnfaip3 | 3360 | 0.136 | 0.2835 | No |
| 51 | Ctsb | 3424 | 0.132 | 0.2828 | No |
| 52 | Gnb2 | 3587 | 0.119 | 0.2730 | No |
| 53 | Casp4 | 3595 | 0.118 | 0.2767 | No |
| 54 | Lipa | 3823 | 0.102 | 0.2606 | No |
| 55 | Vcpip1 | 3906 | 0.096 | 0.2570 | No |
| 56 | Fcer1g | 4146 | 0.077 | 0.2390 | No |
| 57 | Serpinc1 | 4341 | 0.061 | 0.2243 | No |
| 58 | Grb2 | 4392 | 0.057 | 0.2220 | No |
| 59 | Irf2 | 4414 | 0.056 | 0.2222 | No |
| 60 | Lta4h | 4690 | 0.036 | 0.1996 | No |
| 61 | Gmfb | 4798 | 0.027 | 0.1913 | No |
| 62 | Casp1 | 5135 | 0.002 | 0.1622 | No |
| 63 | S100a13 | 5264 | -0.007 | 0.1513 | No |
| 64 | Sh2b3 | 5271 | -0.008 | 0.1511 | No |
| 65 | Lap3 | 5332 | -0.012 | 0.1463 | No |
| 66 | Ppp4c | 5552 | -0.029 | 0.1283 | No |
| 67 | Pik3cg | 5612 | -0.034 | 0.1244 | No |
| 68 | Plscr1 | 5670 | -0.038 | 0.1208 | No |
| 69 | Pfn1 | 6016 | -0.063 | 0.0931 | No |
| 70 | C3 | 6031 | -0.064 | 0.0942 | No |
| 71 | Pla2g4a | 6154 | -0.073 | 0.0863 | No |
| 72 | Dpp4 | 6247 | -0.080 | 0.0812 | No |
| 73 | Rhog | 6502 | -0.102 | 0.0628 | No |
| 74 | Pim1 | 6535 | -0.105 | 0.0638 | No |
| 75 | Usp14 | 6542 | -0.105 | 0.0671 | No |
| 76 | Atox1 | 6575 | -0.108 | 0.0682 | No |
| 77 | Xpnpep1 | 6779 | -0.125 | 0.0551 | No |
| 78 | Dgkg | 6823 | -0.129 | 0.0560 | No |
| 79 | Usp15 | 6854 | -0.132 | 0.0582 | No |
| 80 | Psmb9 | 6927 | -0.137 | 0.0568 | No |
| 81 | Prep | 6985 | -0.141 | 0.0570 | No |
| 82 | Cpq | 7280 | -0.165 | 0.0374 | No |
| 83 | Calm1 | 7347 | -0.170 | 0.0378 | No |
| 84 | Casp3 | 7543 | -0.186 | 0.0276 | No |
| 85 | Stx4a | 7710 | -0.201 | 0.0204 | No |
| 86 | Rce1 | 7713 | -0.201 | 0.0275 | No |
| 87 | Dusp6 | 7763 | -0.205 | 0.0306 | No |
| 88 | Prdm4 | 7892 | -0.217 | 0.0274 | No |
| 89 | Casp7 | 7928 | -0.220 | 0.0323 | No |
| 90 | Car2 | 8000 | -0.226 | 0.0343 | No |
| 91 | Pik3ca | 8032 | -0.228 | 0.0398 | No |
| 92 | Gnai2 | 8171 | -0.239 | 0.0365 | No |
| 93 | Ctsc | 8965 | -0.312 | -0.0212 | No |
| 94 | Kif2a | 9925 | -0.416 | -0.0895 | No |
| 95 | Pik3r5 | 9942 | -0.419 | -0.0758 | No |
| 96 | Cdk5r1 | 9964 | -0.422 | -0.0623 | No |
| 97 | Fyn | 9976 | -0.424 | -0.0480 | No |
| 98 | Ehd1 | 10002 | -0.428 | -0.0347 | No |
| 99 | Zeb1 | 10173 | -0.454 | -0.0330 | No |
| 100 | Akap10 | 10198 | -0.458 | -0.0185 | No |
| 101 | Plaur | 10315 | -0.478 | -0.0114 | No |
| 102 | Prcp | 10316 | -0.479 | 0.0060 | No |
| 103 | Pdp1 | 10447 | -0.496 | 0.0126 | No |
| 104 | Gpd2 | 10468 | -0.499 | 0.0289 | No |
| 105 | Rabif | 10999 | -0.619 | 0.0052 | No |
| 106 | Spock2 | 11049 | -0.635 | 0.0238 | No |
| 107 | Casp9 | 11171 | -0.685 | 0.0381 | No |