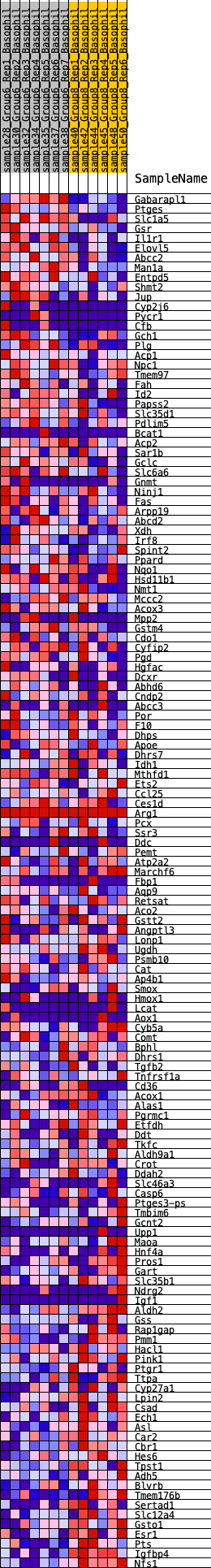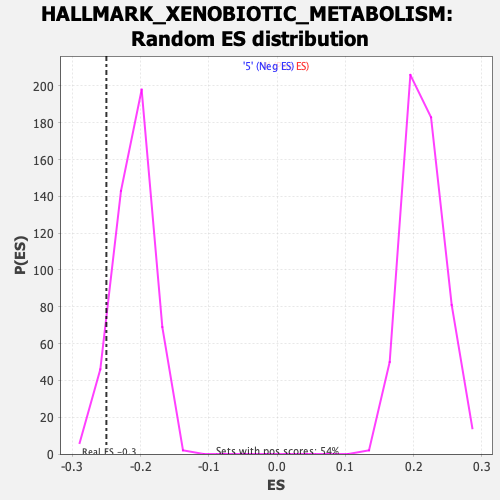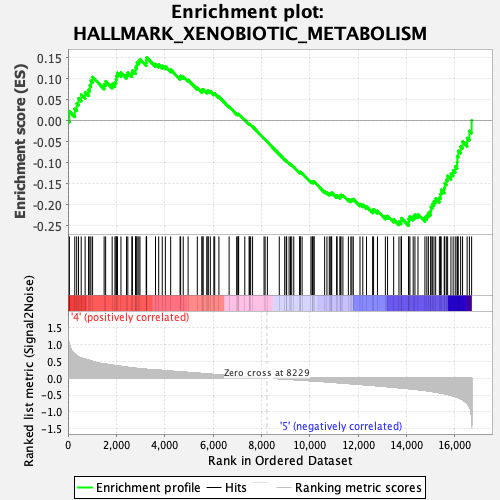
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Basophil.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.2500358 |
| Normalized Enrichment Score (NES) | -1.1846527 |
| Nominal p-value | 0.08405172 |
| FDR q-value | 0.48775122 |
| FWER p-Value | 0.97 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gabarapl1 | 57 | 0.990 | 0.0226 | No |
| 2 | Ptges | 280 | 0.720 | 0.0282 | No |
| 3 | Slc1a5 | 359 | 0.671 | 0.0412 | No |
| 4 | Gsr | 435 | 0.631 | 0.0533 | No |
| 5 | Il1r1 | 546 | 0.600 | 0.0624 | No |
| 6 | Elovl5 | 708 | 0.567 | 0.0676 | No |
| 7 | Abcc2 | 850 | 0.538 | 0.0733 | No |
| 8 | Man1a | 904 | 0.527 | 0.0840 | No |
| 9 | Entpd5 | 948 | 0.516 | 0.0950 | No |
| 10 | Shmt2 | 1017 | 0.498 | 0.1040 | No |
| 11 | Jup | 1494 | 0.425 | 0.0865 | No |
| 12 | Cyp2j6 | 1556 | 0.416 | 0.0937 | No |
| 13 | Pycr1 | 1826 | 0.386 | 0.0877 | No |
| 14 | Cfb | 1942 | 0.377 | 0.0907 | No |
| 15 | Gch1 | 1985 | 0.374 | 0.0980 | No |
| 16 | Plg | 2002 | 0.372 | 0.1068 | No |
| 17 | Acp1 | 2046 | 0.367 | 0.1139 | No |
| 18 | Npc1 | 2190 | 0.352 | 0.1145 | No |
| 19 | Tmem97 | 2421 | 0.326 | 0.1092 | No |
| 20 | Fah | 2474 | 0.321 | 0.1145 | No |
| 21 | Id2 | 2640 | 0.307 | 0.1126 | No |
| 22 | Papss2 | 2663 | 0.306 | 0.1194 | No |
| 23 | Slc35d1 | 2794 | 0.294 | 0.1193 | No |
| 24 | Pdlim5 | 2795 | 0.294 | 0.1270 | No |
| 25 | Bcat1 | 2845 | 0.290 | 0.1317 | No |
| 26 | Acp2 | 2853 | 0.290 | 0.1389 | No |
| 27 | Sar1b | 2928 | 0.283 | 0.1419 | No |
| 28 | Gclc | 2976 | 0.279 | 0.1464 | No |
| 29 | Slc6a6 | 3237 | 0.263 | 0.1376 | No |
| 30 | Gnmt | 3242 | 0.262 | 0.1443 | No |
| 31 | Ninj1 | 3251 | 0.262 | 0.1507 | No |
| 32 | Fas | 3619 | 0.250 | 0.1352 | No |
| 33 | Arpp19 | 3751 | 0.240 | 0.1336 | No |
| 34 | Abcd2 | 3898 | 0.230 | 0.1308 | No |
| 35 | Xdh | 4026 | 0.225 | 0.1291 | No |
| 36 | Irf8 | 4246 | 0.210 | 0.1214 | No |
| 37 | Spint2 | 4634 | 0.189 | 0.1030 | No |
| 38 | Ppard | 4658 | 0.187 | 0.1065 | No |
| 39 | Nqo1 | 4765 | 0.181 | 0.1048 | No |
| 40 | Hsd11b1 | 4967 | 0.169 | 0.0972 | No |
| 41 | Nmt1 | 5347 | 0.149 | 0.0782 | No |
| 42 | Mccc2 | 5542 | 0.138 | 0.0701 | No |
| 43 | Acox3 | 5543 | 0.138 | 0.0738 | No |
| 44 | Mpp2 | 5591 | 0.135 | 0.0745 | No |
| 45 | Gstm4 | 5738 | 0.126 | 0.0690 | No |
| 46 | Cdo1 | 5788 | 0.124 | 0.0693 | No |
| 47 | Cyfip2 | 5798 | 0.123 | 0.0720 | No |
| 48 | Pgd | 5886 | 0.119 | 0.0699 | No |
| 49 | Hgfac | 6032 | 0.110 | 0.0640 | No |
| 50 | Dcxr | 6067 | 0.108 | 0.0648 | No |
| 51 | Abhd6 | 6241 | 0.098 | 0.0569 | No |
| 52 | Cndp2 | 6663 | 0.075 | 0.0335 | No |
| 53 | Abcc3 | 6975 | 0.060 | 0.0163 | No |
| 54 | Por | 7023 | 0.057 | 0.0149 | No |
| 55 | F10 | 7044 | 0.055 | 0.0152 | No |
| 56 | Dhps | 7057 | 0.055 | 0.0159 | No |
| 57 | Apoe | 7311 | 0.042 | 0.0017 | No |
| 58 | Dhrs7 | 7489 | 0.034 | -0.0081 | No |
| 59 | Idh1 | 7512 | 0.033 | -0.0085 | No |
| 60 | Mthfd1 | 7554 | 0.032 | -0.0102 | No |
| 61 | Ets2 | 7633 | 0.028 | -0.0141 | No |
| 62 | Ccl25 | 8107 | 0.004 | -0.0426 | No |
| 63 | Ces1d | 8151 | 0.003 | -0.0451 | No |
| 64 | Arg1 | 8245 | 0.000 | -0.0507 | No |
| 65 | Pcx | 8737 | -0.011 | -0.0801 | No |
| 66 | Ssr3 | 8961 | -0.022 | -0.0930 | No |
| 67 | Ddc | 9024 | -0.025 | -0.0961 | No |
| 68 | Pemt | 9042 | -0.026 | -0.0964 | No |
| 69 | Atp2a2 | 9143 | -0.030 | -0.1017 | No |
| 70 | Marchf6 | 9200 | -0.033 | -0.1042 | No |
| 71 | Fbp1 | 9252 | -0.035 | -0.1063 | No |
| 72 | Aqp9 | 9337 | -0.039 | -0.1104 | No |
| 73 | Retsat | 9581 | -0.050 | -0.1237 | No |
| 74 | Aco2 | 9596 | -0.051 | -0.1232 | No |
| 75 | Gstt2 | 9601 | -0.051 | -0.1221 | No |
| 76 | Angptl3 | 9681 | -0.055 | -0.1254 | No |
| 77 | Lonp1 | 10046 | -0.072 | -0.1455 | No |
| 78 | Ugdh | 10097 | -0.075 | -0.1466 | No |
| 79 | Psmb10 | 10108 | -0.075 | -0.1452 | No |
| 80 | Cat | 10168 | -0.078 | -0.1467 | No |
| 81 | Ap4b1 | 10176 | -0.078 | -0.1451 | No |
| 82 | Smox | 10613 | -0.099 | -0.1688 | No |
| 83 | Hmox1 | 10708 | -0.104 | -0.1717 | No |
| 84 | Lcat | 10804 | -0.110 | -0.1746 | No |
| 85 | Aox1 | 10843 | -0.112 | -0.1739 | No |
| 86 | Cyb5a | 10883 | -0.114 | -0.1733 | No |
| 87 | Comt | 10905 | -0.115 | -0.1715 | No |
| 88 | Bphl | 11109 | -0.127 | -0.1804 | No |
| 89 | Dhrs1 | 11128 | -0.128 | -0.1781 | No |
| 90 | Tgfb2 | 11249 | -0.136 | -0.1818 | No |
| 91 | Tnfrsf1a | 11254 | -0.136 | -0.1785 | No |
| 92 | Cd36 | 11291 | -0.138 | -0.1770 | No |
| 93 | Acox1 | 11368 | -0.142 | -0.1779 | No |
| 94 | Alas1 | 11594 | -0.154 | -0.1874 | No |
| 95 | Pgrmc1 | 11697 | -0.160 | -0.1894 | No |
| 96 | Etfdh | 11741 | -0.162 | -0.1877 | No |
| 97 | Ddt | 11798 | -0.165 | -0.1867 | No |
| 98 | Tkfc | 12075 | -0.181 | -0.1986 | No |
| 99 | Aldh9a1 | 12191 | -0.187 | -0.2007 | No |
| 100 | Crot | 12345 | -0.195 | -0.2048 | No |
| 101 | Ddah2 | 12597 | -0.210 | -0.2144 | No |
| 102 | Slc46a3 | 12637 | -0.213 | -0.2111 | No |
| 103 | Casp6 | 12797 | -0.221 | -0.2149 | No |
| 104 | Ptges3-ps | 13124 | -0.242 | -0.2282 | No |
| 105 | Tmbim6 | 13215 | -0.248 | -0.2271 | No |
| 106 | Gcnt2 | 13467 | -0.265 | -0.2353 | No |
| 107 | Upp1 | 13686 | -0.280 | -0.2411 | No |
| 108 | Maoa | 13773 | -0.283 | -0.2388 | No |
| 109 | Hnf4a | 13791 | -0.285 | -0.2323 | No |
| 110 | Pros1 | 14085 | -0.306 | -0.2420 | Yes |
| 111 | Gart | 14091 | -0.306 | -0.2342 | Yes |
| 112 | Slc35b1 | 14139 | -0.311 | -0.2289 | Yes |
| 113 | Ndrg2 | 14275 | -0.320 | -0.2286 | Yes |
| 114 | Igf1 | 14347 | -0.327 | -0.2242 | Yes |
| 115 | Aldh2 | 14473 | -0.336 | -0.2229 | Yes |
| 116 | Gss | 14760 | -0.360 | -0.2307 | Yes |
| 117 | Rap1gap | 14839 | -0.368 | -0.2257 | Yes |
| 118 | Pmm1 | 14903 | -0.375 | -0.2197 | Yes |
| 119 | Hacl1 | 15000 | -0.386 | -0.2153 | Yes |
| 120 | Pink1 | 15009 | -0.387 | -0.2056 | Yes |
| 121 | Ptgr1 | 15067 | -0.394 | -0.1987 | Yes |
| 122 | Ttpa | 15134 | -0.401 | -0.1921 | Yes |
| 123 | Cyp27a1 | 15204 | -0.409 | -0.1855 | Yes |
| 124 | Lpin2 | 15358 | -0.427 | -0.1835 | Yes |
| 125 | Csad | 15403 | -0.433 | -0.1747 | Yes |
| 126 | Ech1 | 15432 | -0.438 | -0.1649 | Yes |
| 127 | Asl | 15558 | -0.456 | -0.1604 | Yes |
| 128 | Car2 | 15578 | -0.459 | -0.1494 | Yes |
| 129 | Cbr1 | 15645 | -0.471 | -0.1410 | Yes |
| 130 | Hes6 | 15699 | -0.481 | -0.1315 | Yes |
| 131 | Tpst1 | 15836 | -0.503 | -0.1265 | Yes |
| 132 | Adh5 | 15927 | -0.520 | -0.1182 | Yes |
| 133 | Blvrb | 16014 | -0.540 | -0.1092 | Yes |
| 134 | Tmem176b | 16090 | -0.561 | -0.0990 | Yes |
| 135 | Sertad1 | 16102 | -0.564 | -0.0848 | Yes |
| 136 | Slc12a4 | 16137 | -0.572 | -0.0718 | Yes |
| 137 | Gsto1 | 16237 | -0.610 | -0.0617 | Yes |
| 138 | Esr1 | 16317 | -0.637 | -0.0497 | Yes |
| 139 | Pts | 16505 | -0.747 | -0.0413 | Yes |
| 140 | Igfbp4 | 16599 | -0.848 | -0.0246 | Yes |
| 141 | Nfs1 | 16691 | -1.159 | 0.0005 | Yes |