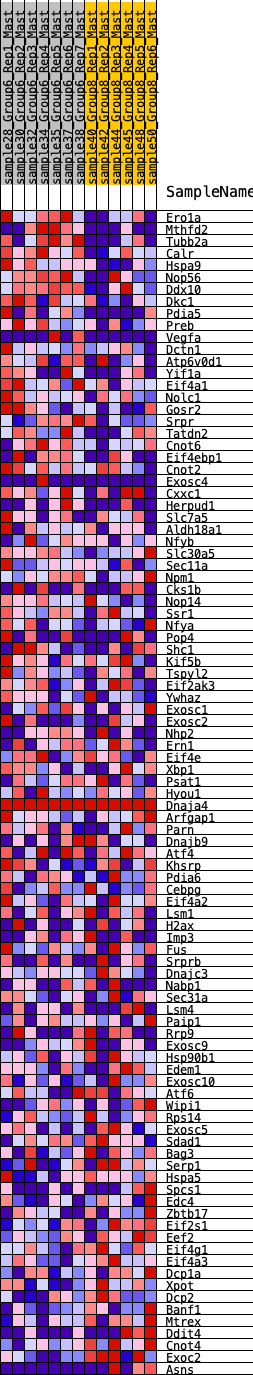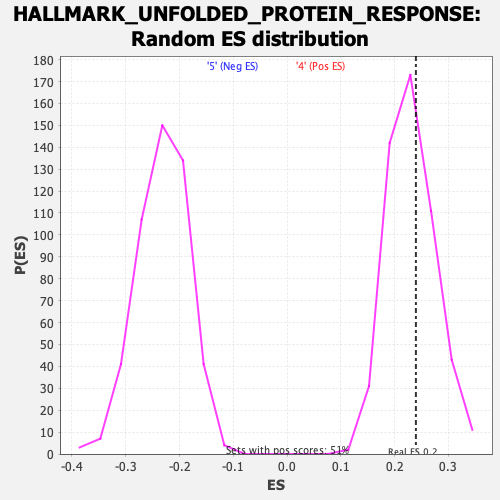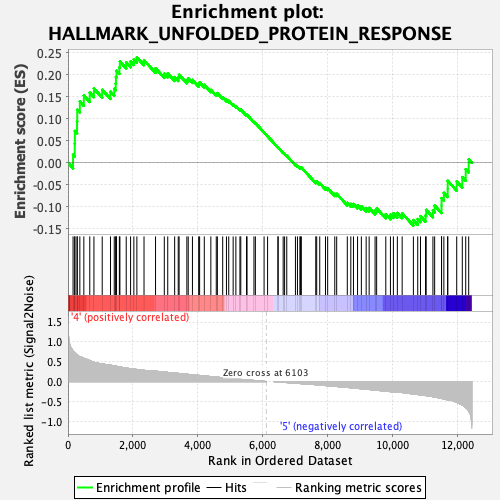
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.23950016 |
| Normalized Enrichment Score (NES) | 1.0386065 |
| Nominal p-value | 0.40350878 |
| FDR q-value | 0.72698027 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ero1a | 156 | 0.773 | 0.0184 | Yes |
| 2 | Mthfd2 | 208 | 0.730 | 0.0435 | Yes |
| 3 | Tubb2a | 212 | 0.726 | 0.0723 | Yes |
| 4 | Calr | 279 | 0.678 | 0.0942 | Yes |
| 5 | Hspa9 | 285 | 0.675 | 0.1208 | Yes |
| 6 | Nop56 | 365 | 0.624 | 0.1394 | Yes |
| 7 | Ddx10 | 488 | 0.584 | 0.1530 | Yes |
| 8 | Dkc1 | 671 | 0.529 | 0.1594 | Yes |
| 9 | Pdia5 | 799 | 0.487 | 0.1686 | Yes |
| 10 | Preb | 1054 | 0.442 | 0.1658 | Yes |
| 11 | Vegfa | 1309 | 0.410 | 0.1616 | Yes |
| 12 | Dctn1 | 1425 | 0.395 | 0.1681 | Yes |
| 13 | Atp6v0d1 | 1470 | 0.388 | 0.1801 | Yes |
| 14 | Yif1a | 1477 | 0.386 | 0.1951 | Yes |
| 15 | Eif4a1 | 1495 | 0.384 | 0.2091 | Yes |
| 16 | Nolc1 | 1583 | 0.369 | 0.2169 | Yes |
| 17 | Gosr2 | 1599 | 0.366 | 0.2303 | Yes |
| 18 | Srpr | 1794 | 0.339 | 0.2282 | Yes |
| 19 | Tatdn2 | 1928 | 0.324 | 0.2304 | Yes |
| 20 | Cnot6 | 2029 | 0.313 | 0.2348 | Yes |
| 21 | Eif4ebp1 | 2122 | 0.302 | 0.2395 | Yes |
| 22 | Cnot2 | 2342 | 0.282 | 0.2331 | No |
| 23 | Exosc4 | 2697 | 0.259 | 0.2148 | No |
| 24 | Cxxc1 | 2967 | 0.239 | 0.2026 | No |
| 25 | Herpud1 | 3073 | 0.229 | 0.2032 | No |
| 26 | Slc7a5 | 3286 | 0.215 | 0.1947 | No |
| 27 | Aldh18a1 | 3396 | 0.205 | 0.1941 | No |
| 28 | Nfyb | 3423 | 0.202 | 0.2001 | No |
| 29 | Slc30a5 | 3662 | 0.182 | 0.1881 | No |
| 30 | Sec11a | 3704 | 0.179 | 0.1920 | No |
| 31 | Npm1 | 3834 | 0.170 | 0.1884 | No |
| 32 | Cks1b | 4026 | 0.155 | 0.1791 | No |
| 33 | Nop14 | 4055 | 0.152 | 0.1829 | No |
| 34 | Ssr1 | 4199 | 0.141 | 0.1770 | No |
| 35 | Nfya | 4401 | 0.124 | 0.1657 | No |
| 36 | Pop4 | 4568 | 0.111 | 0.1567 | No |
| 37 | Shc1 | 4602 | 0.109 | 0.1584 | No |
| 38 | Kif5b | 4766 | 0.096 | 0.1490 | No |
| 39 | Tspyl2 | 4882 | 0.087 | 0.1432 | No |
| 40 | Eif2ak3 | 4952 | 0.082 | 0.1409 | No |
| 41 | Ywhaz | 5088 | 0.072 | 0.1328 | No |
| 42 | Exosc1 | 5172 | 0.067 | 0.1288 | No |
| 43 | Exosc2 | 5297 | 0.056 | 0.1210 | No |
| 44 | Nhp2 | 5323 | 0.054 | 0.1211 | No |
| 45 | Ern1 | 5500 | 0.042 | 0.1085 | No |
| 46 | Eif4e | 5516 | 0.041 | 0.1089 | No |
| 47 | Xbp1 | 5729 | 0.027 | 0.0928 | No |
| 48 | Psat1 | 5773 | 0.023 | 0.0903 | No |
| 49 | Hyou1 | 6041 | 0.005 | 0.0689 | No |
| 50 | Dnaja4 | 6149 | 0.000 | 0.0602 | No |
| 51 | Arfgap1 | 6468 | -0.007 | 0.0347 | No |
| 52 | Parn | 6474 | -0.008 | 0.0346 | No |
| 53 | Dnajb9 | 6629 | -0.018 | 0.0229 | No |
| 54 | Atf4 | 6673 | -0.021 | 0.0202 | No |
| 55 | Khsrp | 6740 | -0.024 | 0.0158 | No |
| 56 | Pdia6 | 7007 | -0.041 | -0.0041 | No |
| 57 | Cebpg | 7069 | -0.044 | -0.0072 | No |
| 58 | Eif4a2 | 7145 | -0.049 | -0.0113 | No |
| 59 | Lsm1 | 7161 | -0.050 | -0.0105 | No |
| 60 | H2ax | 7189 | -0.052 | -0.0107 | No |
| 61 | Imp3 | 7633 | -0.079 | -0.0434 | No |
| 62 | Fus | 7662 | -0.080 | -0.0424 | No |
| 63 | Srprb | 7756 | -0.087 | -0.0465 | No |
| 64 | Dnajc3 | 7933 | -0.100 | -0.0567 | No |
| 65 | Nabp1 | 8000 | -0.104 | -0.0579 | No |
| 66 | Sec31a | 8219 | -0.120 | -0.0708 | No |
| 67 | Lsm4 | 8278 | -0.123 | -0.0705 | No |
| 68 | Paip1 | 8605 | -0.148 | -0.0910 | No |
| 69 | Rrp9 | 8713 | -0.154 | -0.0935 | No |
| 70 | Exosc9 | 8795 | -0.161 | -0.0936 | No |
| 71 | Hsp90b1 | 8921 | -0.172 | -0.0968 | No |
| 72 | Edem1 | 9038 | -0.180 | -0.0990 | No |
| 73 | Exosc10 | 9186 | -0.193 | -0.1032 | No |
| 74 | Atf6 | 9278 | -0.200 | -0.1025 | No |
| 75 | Wipi1 | 9461 | -0.214 | -0.1087 | No |
| 76 | Rps14 | 9509 | -0.218 | -0.1037 | No |
| 77 | Exosc5 | 9791 | -0.244 | -0.1167 | No |
| 78 | Sdad1 | 9934 | -0.253 | -0.1181 | No |
| 79 | Bag3 | 10025 | -0.262 | -0.1149 | No |
| 80 | Serp1 | 10147 | -0.267 | -0.1140 | No |
| 81 | Hspa5 | 10295 | -0.275 | -0.1149 | No |
| 82 | Spcs1 | 10641 | -0.314 | -0.1302 | No |
| 83 | Edc4 | 10775 | -0.326 | -0.1279 | No |
| 84 | Zbtb17 | 10860 | -0.333 | -0.1214 | No |
| 85 | Eif2s1 | 11015 | -0.352 | -0.1198 | No |
| 86 | Eef2 | 11035 | -0.354 | -0.1071 | No |
| 87 | Eif4g1 | 11243 | -0.381 | -0.1086 | No |
| 88 | Eif4a3 | 11296 | -0.388 | -0.0972 | No |
| 89 | Dcp1a | 11508 | -0.424 | -0.0973 | No |
| 90 | Xpot | 11509 | -0.424 | -0.0803 | No |
| 91 | Dcp2 | 11580 | -0.439 | -0.0684 | No |
| 92 | Banf1 | 11698 | -0.459 | -0.0595 | No |
| 93 | Mtrex | 11699 | -0.459 | -0.0411 | No |
| 94 | Ddit4 | 11976 | -0.524 | -0.0424 | No |
| 95 | Cnot4 | 12148 | -0.591 | -0.0326 | No |
| 96 | Exoc2 | 12253 | -0.658 | -0.0147 | No |
| 97 | Asns | 12345 | -0.743 | 0.0078 | No |