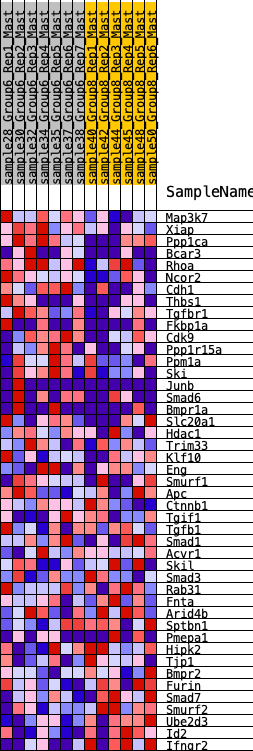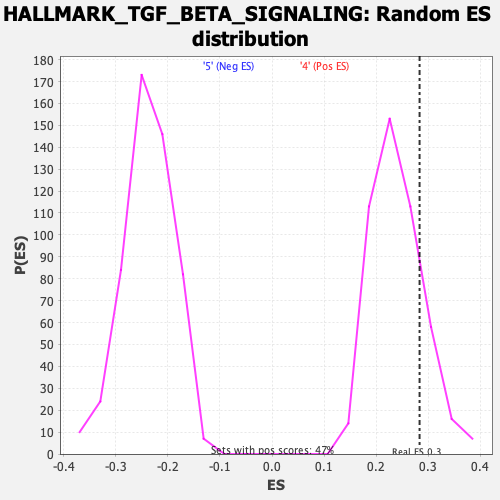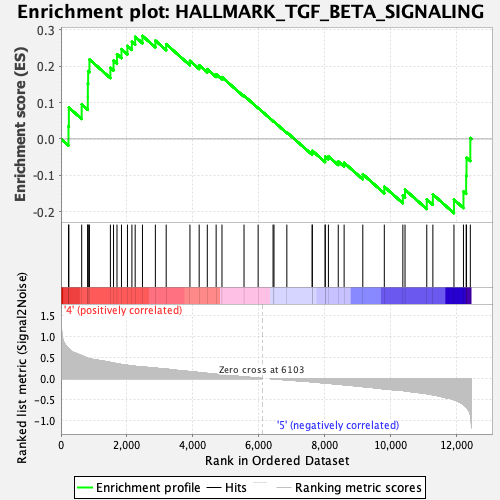
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.2830985 |
| Normalized Enrichment Score (NES) | 1.1815554 |
| Nominal p-value | 0.1814346 |
| FDR q-value | 0.72993094 |
| FWER p-Value | 0.932 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Map3k7 | 227 | 0.716 | 0.0347 | Yes |
| 2 | Xiap | 239 | 0.707 | 0.0861 | Yes |
| 3 | Ppp1ca | 627 | 0.543 | 0.0951 | Yes |
| 4 | Bcar3 | 813 | 0.483 | 0.1159 | Yes |
| 5 | Rhoa | 814 | 0.482 | 0.1516 | Yes |
| 6 | Ncor2 | 823 | 0.480 | 0.1865 | Yes |
| 7 | Cdh1 | 862 | 0.472 | 0.2183 | Yes |
| 8 | Thbs1 | 1497 | 0.383 | 0.1956 | Yes |
| 9 | Tgfbr1 | 1592 | 0.368 | 0.2152 | Yes |
| 10 | Fkbp1a | 1697 | 0.352 | 0.2329 | Yes |
| 11 | Cdk9 | 1833 | 0.335 | 0.2467 | Yes |
| 12 | Ppp1r15a | 2014 | 0.315 | 0.2555 | Yes |
| 13 | Ppm1a | 2149 | 0.299 | 0.2668 | Yes |
| 14 | Ski | 2248 | 0.292 | 0.2806 | Yes |
| 15 | Junb | 2469 | 0.274 | 0.2831 | Yes |
| 16 | Smad6 | 2860 | 0.250 | 0.2702 | No |
| 17 | Bmpr1a | 3188 | 0.224 | 0.2604 | No |
| 18 | Slc20a1 | 3908 | 0.164 | 0.2145 | No |
| 19 | Hdac1 | 4188 | 0.141 | 0.2025 | No |
| 20 | Trim33 | 4434 | 0.122 | 0.1917 | No |
| 21 | Klf10 | 4702 | 0.101 | 0.1777 | No |
| 22 | Eng | 4879 | 0.087 | 0.1699 | No |
| 23 | Smurf1 | 5547 | 0.038 | 0.1190 | No |
| 24 | Apc | 5975 | 0.011 | 0.0853 | No |
| 25 | Ctnnb1 | 6427 | -0.005 | 0.0493 | No |
| 26 | Tgif1 | 6455 | -0.006 | 0.0476 | No |
| 27 | Tgfb1 | 6845 | -0.030 | 0.0184 | No |
| 28 | Smad1 | 7612 | -0.077 | -0.0377 | No |
| 29 | Acvr1 | 7617 | -0.077 | -0.0323 | No |
| 30 | Skil | 8005 | -0.105 | -0.0557 | No |
| 31 | Smad3 | 8009 | -0.105 | -0.0482 | No |
| 32 | Rab31 | 8105 | -0.111 | -0.0476 | No |
| 33 | Fnta | 8403 | -0.133 | -0.0617 | No |
| 34 | Arid4b | 8581 | -0.146 | -0.0652 | No |
| 35 | Sptbn1 | 9147 | -0.190 | -0.0967 | No |
| 36 | Pmepa1 | 9799 | -0.245 | -0.1311 | No |
| 37 | Hipk2 | 10361 | -0.282 | -0.1554 | No |
| 38 | Tjp1 | 10427 | -0.290 | -0.1392 | No |
| 39 | Bmpr2 | 11086 | -0.357 | -0.1658 | No |
| 40 | Furin | 11271 | -0.385 | -0.1522 | No |
| 41 | Smad7 | 11910 | -0.502 | -0.1665 | No |
| 42 | Smurf2 | 12198 | -0.614 | -0.1442 | No |
| 43 | Ube2d3 | 12280 | -0.672 | -0.1010 | No |
| 44 | Id2 | 12291 | -0.680 | -0.0514 | No |
| 45 | Ifngr2 | 12406 | -0.857 | 0.0028 | No |