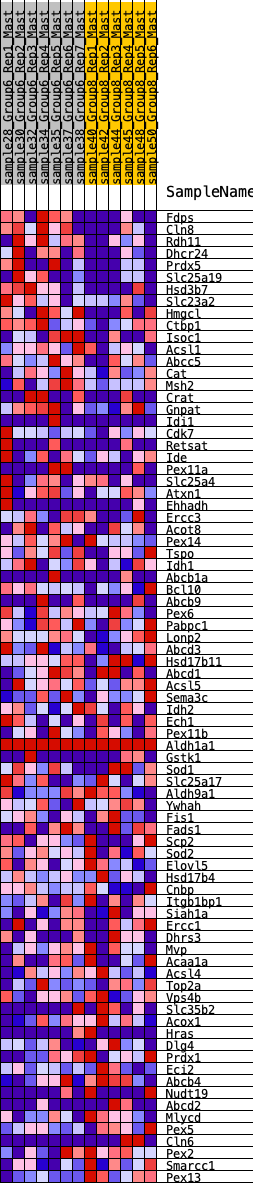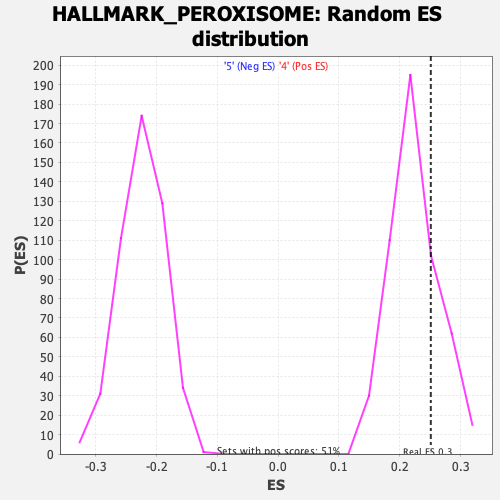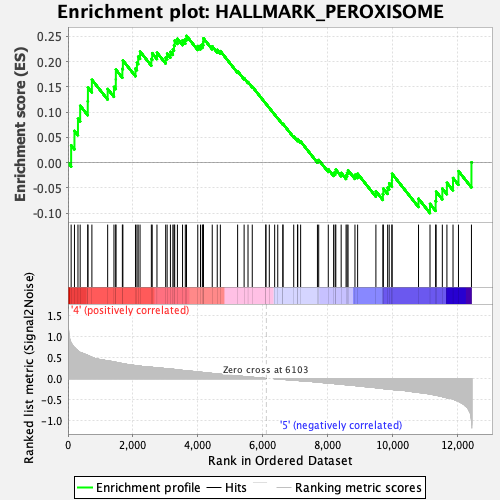
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.25106195 |
| Normalized Enrichment Score (NES) | 1.1195756 |
| Nominal p-value | 0.23735408 |
| FDR q-value | 0.61294556 |
| FWER p-Value | 0.976 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdps | 97 | 0.847 | 0.0345 | Yes |
| 2 | Cln8 | 199 | 0.733 | 0.0631 | Yes |
| 3 | Rdh11 | 307 | 0.655 | 0.0872 | Yes |
| 4 | Dhcr24 | 375 | 0.620 | 0.1128 | Yes |
| 5 | Prdx5 | 609 | 0.552 | 0.1216 | Yes |
| 6 | Slc25a19 | 614 | 0.549 | 0.1487 | Yes |
| 7 | Hsd3b7 | 736 | 0.506 | 0.1642 | Yes |
| 8 | Slc23a2 | 1222 | 0.419 | 0.1460 | Yes |
| 9 | Hmgcl | 1416 | 0.396 | 0.1502 | Yes |
| 10 | Ctbp1 | 1473 | 0.387 | 0.1650 | Yes |
| 11 | Isoc1 | 1475 | 0.387 | 0.1843 | Yes |
| 12 | Acsl1 | 1675 | 0.355 | 0.1860 | Yes |
| 13 | Abcc5 | 1691 | 0.353 | 0.2024 | Yes |
| 14 | Cat | 2079 | 0.308 | 0.1865 | Yes |
| 15 | Msh2 | 2124 | 0.302 | 0.1981 | Yes |
| 16 | Crat | 2162 | 0.298 | 0.2100 | Yes |
| 17 | Gnpat | 2220 | 0.295 | 0.2201 | Yes |
| 18 | Idi1 | 2568 | 0.268 | 0.2055 | Yes |
| 19 | Cdk7 | 2594 | 0.267 | 0.2168 | Yes |
| 20 | Retsat | 2741 | 0.258 | 0.2179 | Yes |
| 21 | Ide | 3009 | 0.235 | 0.2081 | Yes |
| 22 | Pex11a | 3054 | 0.231 | 0.2161 | Yes |
| 23 | Slc25a4 | 3155 | 0.228 | 0.2194 | Yes |
| 24 | Atxn1 | 3231 | 0.221 | 0.2244 | Yes |
| 25 | Ehhadh | 3266 | 0.218 | 0.2325 | Yes |
| 26 | Ercc3 | 3287 | 0.215 | 0.2417 | Yes |
| 27 | Acot8 | 3371 | 0.207 | 0.2453 | Yes |
| 28 | Pex14 | 3529 | 0.193 | 0.2423 | Yes |
| 29 | Tspo | 3614 | 0.186 | 0.2448 | Yes |
| 30 | Idh1 | 3651 | 0.183 | 0.2511 | Yes |
| 31 | Abcb1a | 3998 | 0.158 | 0.2310 | No |
| 32 | Bcl10 | 4082 | 0.150 | 0.2317 | No |
| 33 | Abcb9 | 4140 | 0.146 | 0.2344 | No |
| 34 | Pex6 | 4167 | 0.143 | 0.2395 | No |
| 35 | Pabpc1 | 4172 | 0.143 | 0.2463 | No |
| 36 | Lonp2 | 4444 | 0.121 | 0.2304 | No |
| 37 | Abcd3 | 4598 | 0.109 | 0.2235 | No |
| 38 | Hsd17b11 | 4694 | 0.101 | 0.2209 | No |
| 39 | Abcd1 | 5226 | 0.063 | 0.1811 | No |
| 40 | Acsl5 | 5426 | 0.048 | 0.1674 | No |
| 41 | Sema3c | 5546 | 0.038 | 0.1596 | No |
| 42 | Idh2 | 5677 | 0.031 | 0.1507 | No |
| 43 | Ech1 | 6094 | 0.001 | 0.1170 | No |
| 44 | Pex11b | 6097 | 0.001 | 0.1169 | No |
| 45 | Aldh1a1 | 6202 | 0.000 | 0.1085 | No |
| 46 | Gstk1 | 6367 | -0.002 | 0.0953 | No |
| 47 | Sod1 | 6462 | -0.007 | 0.0880 | No |
| 48 | Slc25a17 | 6621 | -0.017 | 0.0761 | No |
| 49 | Aldh9a1 | 6622 | -0.017 | 0.0770 | No |
| 50 | Ywhah | 6954 | -0.037 | 0.0520 | No |
| 51 | Fis1 | 7070 | -0.044 | 0.0449 | No |
| 52 | Fads1 | 7079 | -0.045 | 0.0465 | No |
| 53 | Scp2 | 7167 | -0.050 | 0.0420 | No |
| 54 | Sod2 | 7685 | -0.082 | 0.0043 | No |
| 55 | Elovl5 | 7724 | -0.085 | 0.0055 | No |
| 56 | Hsd17b4 | 8019 | -0.106 | -0.0130 | No |
| 57 | Cnbp | 8183 | -0.117 | -0.0204 | No |
| 58 | Itgb1bp1 | 8230 | -0.120 | -0.0180 | No |
| 59 | Siah1a | 8251 | -0.122 | -0.0136 | No |
| 60 | Ercc1 | 8416 | -0.134 | -0.0201 | No |
| 61 | Dhrs3 | 8567 | -0.145 | -0.0250 | No |
| 62 | Mvp | 8598 | -0.148 | -0.0200 | No |
| 63 | Acaa1a | 8628 | -0.149 | -0.0149 | No |
| 64 | Acsl4 | 8841 | -0.165 | -0.0238 | No |
| 65 | Top2a | 8923 | -0.172 | -0.0218 | No |
| 66 | Vps4b | 9486 | -0.217 | -0.0564 | No |
| 67 | Slc35b2 | 9701 | -0.236 | -0.0619 | No |
| 68 | Acox1 | 9715 | -0.237 | -0.0511 | No |
| 69 | Hras | 9848 | -0.248 | -0.0493 | No |
| 70 | Dlg4 | 9896 | -0.250 | -0.0406 | No |
| 71 | Prdx1 | 9980 | -0.257 | -0.0345 | No |
| 72 | Eci2 | 9983 | -0.257 | -0.0217 | No |
| 73 | Abcb4 | 10799 | -0.329 | -0.0712 | No |
| 74 | Nudt19 | 11153 | -0.367 | -0.0814 | No |
| 75 | Abcd2 | 11326 | -0.392 | -0.0757 | No |
| 76 | Mlycd | 11341 | -0.394 | -0.0571 | No |
| 77 | Pex5 | 11533 | -0.429 | -0.0510 | No |
| 78 | Cln6 | 11674 | -0.455 | -0.0396 | No |
| 79 | Pex2 | 11862 | -0.490 | -0.0302 | No |
| 80 | Smarcc1 | 12030 | -0.542 | -0.0166 | No |
| 81 | Pex13 | 12430 | -0.994 | 0.0009 | No |