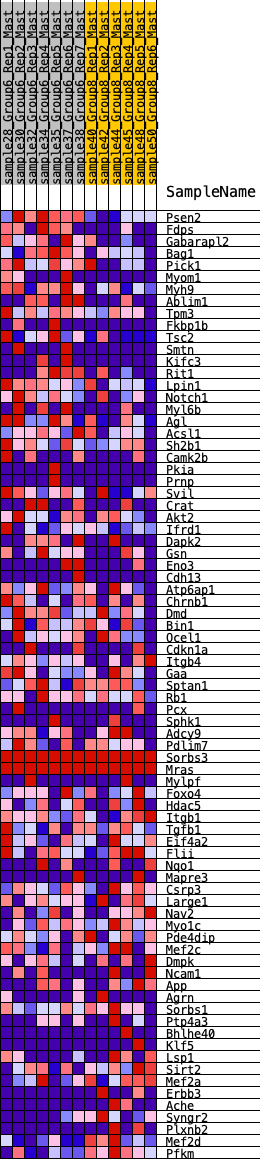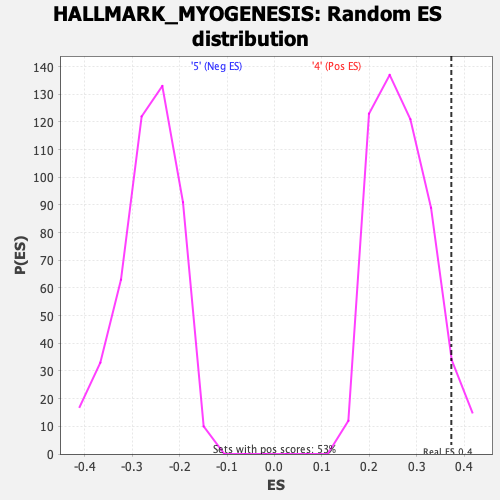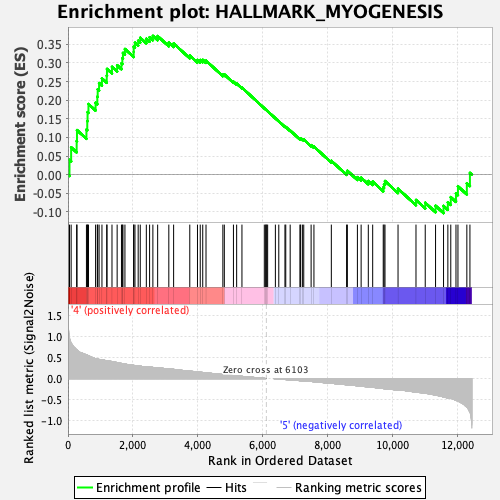
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.3727388 |
| Normalized Enrichment Score (NES) | 1.3866585 |
| Nominal p-value | 0.03954802 |
| FDR q-value | 0.5882917 |
| FWER p-Value | 0.567 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Psen2 | 46 | 0.977 | 0.0397 | Yes |
| 2 | Fdps | 97 | 0.847 | 0.0733 | Yes |
| 3 | Gabarapl2 | 267 | 0.685 | 0.0901 | Yes |
| 4 | Bag1 | 277 | 0.679 | 0.1196 | Yes |
| 5 | Pick1 | 568 | 0.562 | 0.1212 | Yes |
| 6 | Myom1 | 594 | 0.555 | 0.1438 | Yes |
| 7 | Myh9 | 603 | 0.553 | 0.1678 | Yes |
| 8 | Ablim1 | 631 | 0.542 | 0.1897 | Yes |
| 9 | Tpm3 | 850 | 0.474 | 0.1931 | Yes |
| 10 | Fkbp1b | 903 | 0.462 | 0.2095 | Yes |
| 11 | Tsc2 | 915 | 0.461 | 0.2291 | Yes |
| 12 | Smtn | 958 | 0.456 | 0.2460 | Yes |
| 13 | Kifc3 | 1046 | 0.443 | 0.2586 | Yes |
| 14 | Rit1 | 1194 | 0.424 | 0.2656 | Yes |
| 15 | Lpin1 | 1201 | 0.423 | 0.2840 | Yes |
| 16 | Notch1 | 1355 | 0.406 | 0.2896 | Yes |
| 17 | Myl6b | 1514 | 0.381 | 0.2938 | Yes |
| 18 | Agl | 1647 | 0.359 | 0.2991 | Yes |
| 19 | Acsl1 | 1675 | 0.355 | 0.3127 | Yes |
| 20 | Sh2b1 | 1693 | 0.353 | 0.3270 | Yes |
| 21 | Camk2b | 1754 | 0.345 | 0.3375 | Yes |
| 22 | Pkia | 2023 | 0.314 | 0.3298 | Yes |
| 23 | Prnp | 2025 | 0.314 | 0.3436 | Yes |
| 24 | Svil | 2063 | 0.310 | 0.3544 | Yes |
| 25 | Crat | 2162 | 0.298 | 0.3597 | Yes |
| 26 | Akt2 | 2226 | 0.295 | 0.3677 | Yes |
| 27 | Ifrd1 | 2413 | 0.278 | 0.3651 | Yes |
| 28 | Dapk2 | 2514 | 0.271 | 0.3690 | Yes |
| 29 | Gsn | 2615 | 0.265 | 0.3727 | Yes |
| 30 | Eno3 | 2762 | 0.255 | 0.3723 | No |
| 31 | Cdh13 | 3106 | 0.228 | 0.3547 | No |
| 32 | Atp6ap1 | 3254 | 0.219 | 0.3525 | No |
| 33 | Chrnb1 | 3752 | 0.176 | 0.3201 | No |
| 34 | Dmd | 3989 | 0.158 | 0.3081 | No |
| 35 | Bin1 | 4074 | 0.150 | 0.3080 | No |
| 36 | Ocel1 | 4151 | 0.145 | 0.3083 | No |
| 37 | Cdkn1a | 4252 | 0.137 | 0.3063 | No |
| 38 | Itgb4 | 4771 | 0.095 | 0.2686 | No |
| 39 | Gaa | 4816 | 0.093 | 0.2692 | No |
| 40 | Sptan1 | 5097 | 0.071 | 0.2497 | No |
| 41 | Rb1 | 5196 | 0.065 | 0.2446 | No |
| 42 | Pcx | 5359 | 0.052 | 0.2338 | No |
| 43 | Sphk1 | 6048 | 0.005 | 0.1784 | No |
| 44 | Adcy9 | 6091 | 0.001 | 0.1750 | No |
| 45 | Pdlim7 | 6100 | 0.000 | 0.1744 | No |
| 46 | Sorbs3 | 6122 | 0.000 | 0.1727 | No |
| 47 | Mras | 6151 | 0.000 | 0.1704 | No |
| 48 | Mylpf | 6389 | -0.003 | 0.1514 | No |
| 49 | Foxo4 | 6494 | -0.009 | 0.1434 | No |
| 50 | Hdac5 | 6684 | -0.021 | 0.1290 | No |
| 51 | Itgb1 | 6709 | -0.023 | 0.1281 | No |
| 52 | Tgfb1 | 6845 | -0.030 | 0.1185 | No |
| 53 | Eif4a2 | 7145 | -0.049 | 0.0965 | No |
| 54 | Flii | 7166 | -0.050 | 0.0972 | No |
| 55 | Nqo1 | 7221 | -0.054 | 0.0952 | No |
| 56 | Mapre3 | 7263 | -0.057 | 0.0944 | No |
| 57 | Csrp3 | 7490 | -0.068 | 0.0792 | No |
| 58 | Large1 | 7577 | -0.075 | 0.0755 | No |
| 59 | Nav2 | 8113 | -0.112 | 0.0372 | No |
| 60 | Myo1c | 8583 | -0.146 | 0.0058 | No |
| 61 | Pde4dip | 8607 | -0.148 | 0.0105 | No |
| 62 | Mef2c | 8916 | -0.171 | -0.0068 | No |
| 63 | Dmpk | 9031 | -0.180 | -0.0080 | No |
| 64 | Ncam1 | 9252 | -0.197 | -0.0170 | No |
| 65 | App | 9387 | -0.207 | -0.0187 | No |
| 66 | Agrn | 9712 | -0.237 | -0.0343 | No |
| 67 | Sorbs1 | 9737 | -0.240 | -0.0256 | No |
| 68 | Ptp4a3 | 9766 | -0.242 | -0.0171 | No |
| 69 | Bhlhe40 | 10168 | -0.267 | -0.0377 | No |
| 70 | Klf5 | 10721 | -0.322 | -0.0680 | No |
| 71 | Lsp1 | 11006 | -0.350 | -0.0754 | No |
| 72 | Sirt2 | 11324 | -0.392 | -0.0836 | No |
| 73 | Mef2a | 11570 | -0.436 | -0.0840 | No |
| 74 | Erbb3 | 11703 | -0.460 | -0.0742 | No |
| 75 | Ache | 11793 | -0.474 | -0.0604 | No |
| 76 | Syngr2 | 11953 | -0.515 | -0.0503 | No |
| 77 | Plxnb2 | 12014 | -0.538 | -0.0313 | No |
| 78 | Mef2d | 12288 | -0.678 | -0.0232 | No |
| 79 | Pfkm | 12383 | -0.798 | 0.0047 | No |