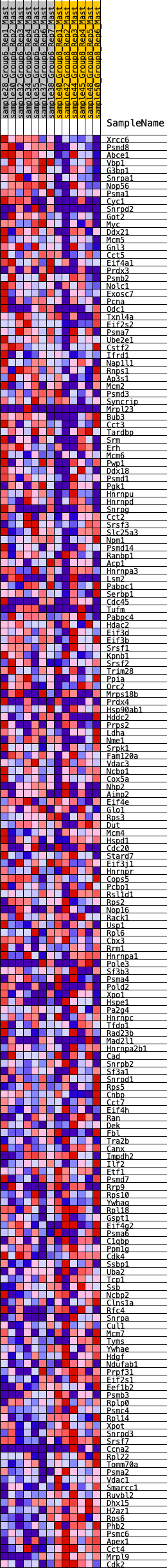
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
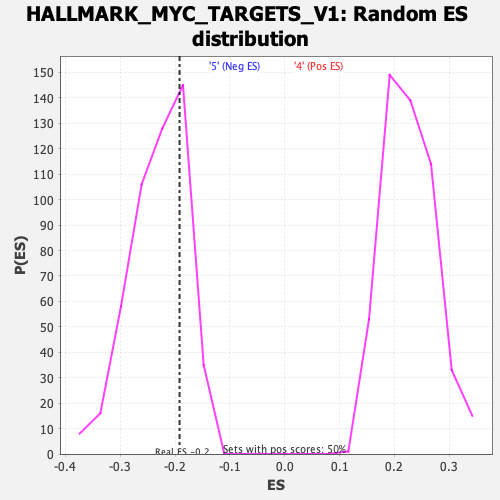
| GeneSet | HALLMARK_MYC_TARGETS_V1 |
| Enrichment Score (ES) | -0.19188808 |
| Normalized Enrichment Score (NES) | -0.8351369 |
| Nominal p-value | 0.7419355 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xrcc6 | 82 | 0.870 | 0.0124 | No |
| 2 | Psmd8 | 146 | 0.786 | 0.0246 | No |
| 3 | Abce1 | 162 | 0.769 | 0.0403 | No |
| 4 | Vbp1 | 203 | 0.731 | 0.0530 | No |
| 5 | G3bp1 | 295 | 0.666 | 0.0603 | No |
| 6 | Snrpa1 | 317 | 0.648 | 0.0728 | No |
| 7 | Nop56 | 365 | 0.624 | 0.0827 | No |
| 8 | Psma1 | 553 | 0.569 | 0.0799 | No |
| 9 | Cyc1 | 566 | 0.565 | 0.0913 | No |
| 10 | Snrpd2 | 625 | 0.545 | 0.0986 | No |
| 11 | Got2 | 643 | 0.537 | 0.1090 | No |
| 12 | Myc | 692 | 0.522 | 0.1165 | No |
| 13 | Ddx21 | 1202 | 0.423 | 0.0843 | No |
| 14 | Mcm5 | 1401 | 0.400 | 0.0769 | No |
| 15 | Gnl3 | 1422 | 0.395 | 0.0840 | No |
| 16 | Cct5 | 1474 | 0.387 | 0.0883 | No |
| 17 | Eif4a1 | 1495 | 0.384 | 0.0951 | No |
| 18 | Prdx3 | 1552 | 0.374 | 0.0988 | No |
| 19 | Psmb2 | 1573 | 0.371 | 0.1053 | No |
| 20 | Nolc1 | 1583 | 0.369 | 0.1127 | No |
| 21 | Exosc7 | 1613 | 0.364 | 0.1183 | No |
| 22 | Pcna | 1792 | 0.339 | 0.1112 | No |
| 23 | Odc1 | 2042 | 0.312 | 0.0978 | No |
| 24 | Txnl4a | 2058 | 0.310 | 0.1034 | No |
| 25 | Eif2s2 | 2101 | 0.306 | 0.1066 | No |
| 26 | Psma7 | 2167 | 0.297 | 0.1079 | No |
| 27 | Ube2e1 | 2211 | 0.297 | 0.1109 | No |
| 28 | Cstf2 | 2232 | 0.294 | 0.1157 | No |
| 29 | Ifrd1 | 2413 | 0.278 | 0.1071 | No |
| 30 | Nap1l1 | 2498 | 0.274 | 0.1063 | No |
| 31 | Rnps1 | 2524 | 0.270 | 0.1102 | No |
| 32 | Ap3s1 | 2634 | 0.263 | 0.1071 | No |
| 33 | Mcm2 | 2663 | 0.260 | 0.1105 | No |
| 34 | Psmd3 | 2667 | 0.260 | 0.1160 | No |
| 35 | Syncrip | 2679 | 0.259 | 0.1208 | No |
| 36 | Mrpl23 | 2699 | 0.259 | 0.1249 | No |
| 37 | Bub3 | 2864 | 0.250 | 0.1170 | No |
| 38 | Cct3 | 2878 | 0.248 | 0.1214 | No |
| 39 | Tardbp | 2974 | 0.239 | 0.1189 | No |
| 40 | Srm | 2980 | 0.238 | 0.1237 | No |
| 41 | Erh | 3033 | 0.233 | 0.1246 | No |
| 42 | Mcm6 | 3069 | 0.230 | 0.1268 | No |
| 43 | Pwp1 | 3081 | 0.229 | 0.1309 | No |
| 44 | Ddx18 | 3244 | 0.220 | 0.1225 | No |
| 45 | Psmd1 | 3250 | 0.219 | 0.1269 | No |
| 46 | Pgk1 | 3267 | 0.218 | 0.1304 | No |
| 47 | Hnrnpu | 3410 | 0.204 | 0.1233 | No |
| 48 | Hnrnpd | 3490 | 0.197 | 0.1212 | No |
| 49 | Snrpg | 3531 | 0.193 | 0.1221 | No |
| 50 | Cct2 | 3571 | 0.189 | 0.1231 | No |
| 51 | Srsf3 | 3748 | 0.177 | 0.1126 | No |
| 52 | Slc25a3 | 3763 | 0.175 | 0.1153 | No |
| 53 | Npm1 | 3834 | 0.170 | 0.1134 | No |
| 54 | Psmd14 | 3871 | 0.168 | 0.1141 | No |
| 55 | Ranbp1 | 4035 | 0.154 | 0.1042 | No |
| 56 | Acp1 | 4046 | 0.153 | 0.1067 | No |
| 57 | Hnrnpa3 | 4079 | 0.150 | 0.1074 | No |
| 58 | Lsm2 | 4091 | 0.149 | 0.1098 | No |
| 59 | Pabpc1 | 4172 | 0.143 | 0.1064 | No |
| 60 | Serbp1 | 4187 | 0.141 | 0.1084 | No |
| 61 | Cdc45 | 4192 | 0.141 | 0.1111 | No |
| 62 | Tufm | 4238 | 0.138 | 0.1105 | No |
| 63 | Pabpc4 | 4280 | 0.135 | 0.1101 | No |
| 64 | Hdac2 | 4370 | 0.127 | 0.1056 | No |
| 65 | Eif3d | 4397 | 0.124 | 0.1063 | No |
| 66 | Eif3b | 4402 | 0.124 | 0.1086 | No |
| 67 | Srsf1 | 4469 | 0.119 | 0.1059 | No |
| 68 | Kpnb1 | 4475 | 0.118 | 0.1081 | No |
| 69 | Srsf2 | 4519 | 0.115 | 0.1071 | No |
| 70 | Trim28 | 4531 | 0.114 | 0.1087 | No |
| 71 | Ppia | 4648 | 0.105 | 0.1015 | No |
| 72 | Orc2 | 4651 | 0.105 | 0.1037 | No |
| 73 | Mrps18b | 4765 | 0.096 | 0.0966 | No |
| 74 | Prdx4 | 4851 | 0.090 | 0.0916 | No |
| 75 | Hsp90ab1 | 5035 | 0.076 | 0.0783 | No |
| 76 | Hddc2 | 5042 | 0.075 | 0.0795 | No |
| 77 | Prps2 | 5110 | 0.070 | 0.0756 | No |
| 78 | Ldha | 5127 | 0.069 | 0.0758 | No |
| 79 | Nme1 | 5166 | 0.067 | 0.0741 | No |
| 80 | Srpk1 | 5192 | 0.065 | 0.0735 | No |
| 81 | Fam120a | 5206 | 0.064 | 0.0739 | No |
| 82 | Vdac3 | 5243 | 0.061 | 0.0723 | No |
| 83 | Ncbp1 | 5286 | 0.057 | 0.0701 | No |
| 84 | Cox5a | 5287 | 0.057 | 0.0714 | No |
| 85 | Nhp2 | 5323 | 0.054 | 0.0697 | No |
| 86 | Aimp2 | 5347 | 0.052 | 0.0690 | No |
| 87 | Eif4e | 5516 | 0.041 | 0.0561 | No |
| 88 | Glo1 | 5527 | 0.040 | 0.0562 | No |
| 89 | Rps3 | 5611 | 0.035 | 0.0502 | No |
| 90 | Dut | 5622 | 0.035 | 0.0501 | No |
| 91 | Mcm4 | 5834 | 0.018 | 0.0333 | No |
| 92 | Hspd1 | 5886 | 0.015 | 0.0295 | No |
| 93 | Cdc20 | 6019 | 0.007 | 0.0189 | No |
| 94 | Stard7 | 6068 | 0.003 | 0.0150 | No |
| 95 | Eif3j1 | 6524 | -0.011 | -0.0219 | No |
| 96 | Hnrnpr | 6558 | -0.013 | -0.0243 | No |
| 97 | Cops5 | 6565 | -0.013 | -0.0245 | No |
| 98 | Pcbp1 | 6609 | -0.016 | -0.0276 | No |
| 99 | Rsl1d1 | 6614 | -0.016 | -0.0276 | No |
| 100 | Rps2 | 6625 | -0.017 | -0.0280 | No |
| 101 | Nop16 | 6665 | -0.020 | -0.0307 | No |
| 102 | Rack1 | 6723 | -0.024 | -0.0349 | No |
| 103 | Usp1 | 6796 | -0.027 | -0.0401 | No |
| 104 | Rpl6 | 6799 | -0.028 | -0.0397 | No |
| 105 | Cbx3 | 6912 | -0.034 | -0.0481 | No |
| 106 | Rrm1 | 7024 | -0.042 | -0.0562 | No |
| 107 | Hnrnpa1 | 7076 | -0.044 | -0.0594 | No |
| 108 | Pole3 | 7130 | -0.048 | -0.0627 | No |
| 109 | Sf3b3 | 7142 | -0.049 | -0.0625 | No |
| 110 | Psma4 | 7203 | -0.053 | -0.0662 | No |
| 111 | Pold2 | 7364 | -0.060 | -0.0780 | No |
| 112 | Xpo1 | 7466 | -0.067 | -0.0847 | No |
| 113 | Hspe1 | 7547 | -0.072 | -0.0897 | No |
| 114 | Pa2g4 | 7584 | -0.075 | -0.0910 | No |
| 115 | Hnrnpc | 7619 | -0.078 | -0.0920 | No |
| 116 | Tfdp1 | 7668 | -0.081 | -0.0942 | No |
| 117 | Rad23b | 7791 | -0.089 | -0.1022 | No |
| 118 | Mad2l1 | 7837 | -0.093 | -0.1038 | No |
| 119 | Hnrnpa2b1 | 7844 | -0.094 | -0.1022 | No |
| 120 | Cad | 8021 | -0.106 | -0.1143 | No |
| 121 | Snrpb2 | 8051 | -0.108 | -0.1143 | No |
| 122 | Sf3a1 | 8076 | -0.109 | -0.1138 | No |
| 123 | Snrpd1 | 8086 | -0.110 | -0.1121 | No |
| 124 | Rps5 | 8123 | -0.112 | -0.1126 | No |
| 125 | Cnbp | 8183 | -0.117 | -0.1149 | No |
| 126 | Cct7 | 8211 | -0.119 | -0.1145 | No |
| 127 | Eif4h | 8277 | -0.123 | -0.1170 | No |
| 128 | Ran | 8311 | -0.126 | -0.1170 | No |
| 129 | Dek | 8346 | -0.129 | -0.1169 | No |
| 130 | Fbl | 8357 | -0.130 | -0.1149 | No |
| 131 | Tra2b | 8392 | -0.132 | -0.1147 | No |
| 132 | Canx | 8412 | -0.134 | -0.1133 | No |
| 133 | Impdh2 | 8427 | -0.135 | -0.1115 | No |
| 134 | Ilf2 | 8437 | -0.136 | -0.1093 | No |
| 135 | Etf1 | 8527 | -0.142 | -0.1134 | No |
| 136 | Psmd7 | 8687 | -0.153 | -0.1230 | No |
| 137 | Rrp9 | 8713 | -0.154 | -0.1217 | No |
| 138 | Rps10 | 8741 | -0.157 | -0.1204 | No |
| 139 | Ywhaq | 8852 | -0.166 | -0.1257 | No |
| 140 | Rpl18 | 8873 | -0.168 | -0.1237 | No |
| 141 | Gspt1 | 8907 | -0.170 | -0.1226 | No |
| 142 | Eif4g2 | 8940 | -0.173 | -0.1214 | No |
| 143 | Psma6 | 9094 | -0.185 | -0.1299 | No |
| 144 | C1qbp | 9203 | -0.194 | -0.1344 | No |
| 145 | Ppm1g | 9283 | -0.200 | -0.1365 | No |
| 146 | Cdk4 | 9360 | -0.206 | -0.1381 | No |
| 147 | Ssbp1 | 9424 | -0.209 | -0.1387 | No |
| 148 | Uba2 | 9706 | -0.236 | -0.1564 | No |
| 149 | Tcp1 | 9811 | -0.245 | -0.1595 | No |
| 150 | Ssb | 9990 | -0.258 | -0.1684 | No |
| 151 | Ncbp2 | 10005 | -0.260 | -0.1638 | No |
| 152 | Clns1a | 10013 | -0.260 | -0.1587 | No |
| 153 | Rfc4 | 10035 | -0.263 | -0.1546 | No |
| 154 | Snrpa | 10289 | -0.274 | -0.1692 | No |
| 155 | Cul1 | 10371 | -0.284 | -0.1696 | No |
| 156 | Mcm7 | 10434 | -0.291 | -0.1682 | No |
| 157 | Tyms | 10436 | -0.291 | -0.1619 | No |
| 158 | Ywhae | 10731 | -0.322 | -0.1788 | No |
| 159 | Hdgf | 10892 | -0.338 | -0.1845 | Yes |
| 160 | Ndufab1 | 10894 | -0.339 | -0.1771 | Yes |
| 161 | Prpf31 | 10920 | -0.342 | -0.1716 | Yes |
| 162 | Eif2s1 | 11015 | -0.352 | -0.1716 | Yes |
| 163 | Eef1b2 | 11124 | -0.364 | -0.1724 | Yes |
| 164 | Psmb3 | 11235 | -0.380 | -0.1730 | Yes |
| 165 | Rplp0 | 11267 | -0.385 | -0.1671 | Yes |
| 166 | Psmc4 | 11448 | -0.412 | -0.1727 | Yes |
| 167 | Rpl14 | 11467 | -0.416 | -0.1651 | Yes |
| 168 | Xpot | 11509 | -0.424 | -0.1591 | Yes |
| 169 | Snrpd3 | 11522 | -0.427 | -0.1507 | Yes |
| 170 | Srsf7 | 11554 | -0.433 | -0.1437 | Yes |
| 171 | Ccna2 | 11559 | -0.434 | -0.1345 | Yes |
| 172 | Rpl22 | 11573 | -0.437 | -0.1260 | Yes |
| 173 | Tomm70a | 11581 | -0.439 | -0.1169 | Yes |
| 174 | Psma2 | 11717 | -0.463 | -0.1177 | Yes |
| 175 | Vdac1 | 12018 | -0.539 | -0.1304 | Yes |
| 176 | Smarcc1 | 12030 | -0.542 | -0.1194 | Yes |
| 177 | Ruvbl2 | 12093 | -0.568 | -0.1119 | Yes |
| 178 | Dhx15 | 12118 | -0.578 | -0.1012 | Yes |
| 179 | H2az1 | 12120 | -0.579 | -0.0886 | Yes |
| 180 | Rps6 | 12153 | -0.594 | -0.0781 | Yes |
| 181 | Phb2 | 12285 | -0.677 | -0.0739 | Yes |
| 182 | Psmc6 | 12346 | -0.744 | -0.0625 | Yes |
| 183 | Apex1 | 12371 | -0.780 | -0.0473 | Yes |
| 184 | Cct4 | 12372 | -0.781 | -0.0302 | Yes |
| 185 | Mrpl9 | 12389 | -0.808 | -0.0137 | Yes |
| 186 | Cdk2 | 12393 | -0.813 | 0.0039 | Yes |