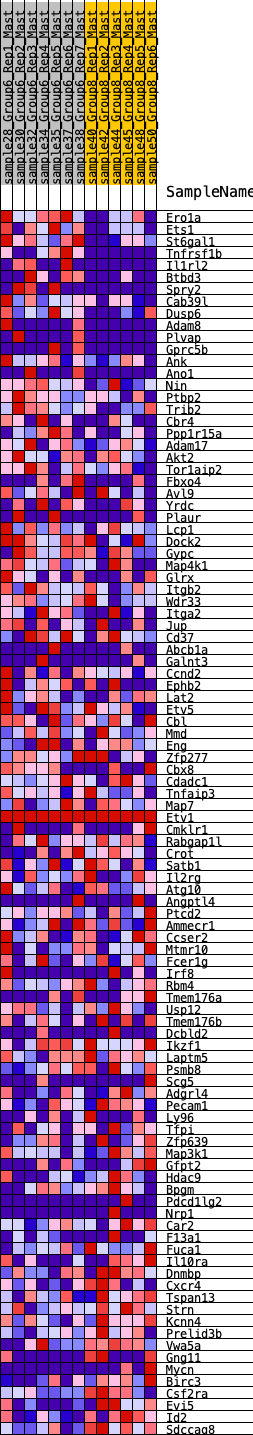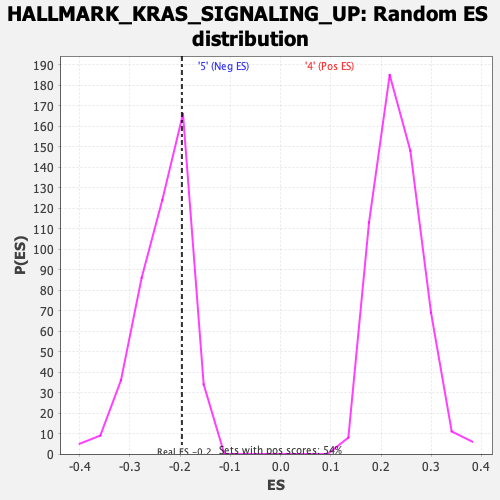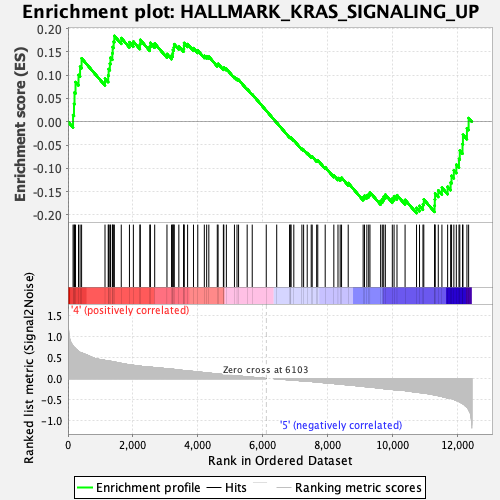
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | -0.19626226 |
| Normalized Enrichment Score (NES) | -0.8418317 |
| Nominal p-value | 0.7695652 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ero1a | 156 | 0.773 | 0.0145 | No |
| 2 | Ets1 | 185 | 0.747 | 0.0384 | No |
| 3 | St6gal1 | 201 | 0.732 | 0.0628 | No |
| 4 | Tnfrsf1b | 232 | 0.709 | 0.0852 | No |
| 5 | Il1rl2 | 324 | 0.643 | 0.1004 | No |
| 6 | Btbd3 | 372 | 0.621 | 0.1184 | No |
| 7 | Spry2 | 419 | 0.613 | 0.1361 | No |
| 8 | Cab39l | 1140 | 0.431 | 0.0929 | No |
| 9 | Dusp6 | 1239 | 0.417 | 0.0995 | No |
| 10 | Adam8 | 1252 | 0.414 | 0.1131 | No |
| 11 | Plvap | 1290 | 0.412 | 0.1245 | No |
| 12 | Gprc5b | 1308 | 0.410 | 0.1375 | No |
| 13 | Ank | 1366 | 0.404 | 0.1471 | No |
| 14 | Ano1 | 1376 | 0.403 | 0.1605 | No |
| 15 | Nin | 1407 | 0.398 | 0.1720 | No |
| 16 | Ptbp2 | 1427 | 0.394 | 0.1843 | No |
| 17 | Trib2 | 1641 | 0.361 | 0.1797 | No |
| 18 | Cbr4 | 1893 | 0.327 | 0.1708 | No |
| 19 | Ppp1r15a | 2014 | 0.315 | 0.1721 | No |
| 20 | Adam17 | 2216 | 0.296 | 0.1662 | No |
| 21 | Akt2 | 2226 | 0.295 | 0.1758 | No |
| 22 | Tor1aip2 | 2520 | 0.270 | 0.1615 | No |
| 23 | Fbxo4 | 2539 | 0.268 | 0.1695 | No |
| 24 | Avl9 | 2671 | 0.260 | 0.1679 | No |
| 25 | Yrdc | 3049 | 0.232 | 0.1455 | No |
| 26 | Plaur | 3190 | 0.224 | 0.1420 | No |
| 27 | Lcp1 | 3227 | 0.221 | 0.1468 | No |
| 28 | Dock2 | 3232 | 0.220 | 0.1542 | No |
| 29 | Gypc | 3261 | 0.218 | 0.1596 | No |
| 30 | Map4k1 | 3271 | 0.217 | 0.1665 | No |
| 31 | Glrx | 3415 | 0.203 | 0.1620 | No |
| 32 | Itgb2 | 3561 | 0.190 | 0.1570 | No |
| 33 | Wdr33 | 3573 | 0.189 | 0.1627 | No |
| 34 | Itga2 | 3576 | 0.189 | 0.1691 | No |
| 35 | Jup | 3685 | 0.180 | 0.1667 | No |
| 36 | Cd37 | 3866 | 0.168 | 0.1580 | No |
| 37 | Abcb1a | 3998 | 0.158 | 0.1529 | No |
| 38 | Galnt3 | 4200 | 0.141 | 0.1416 | No |
| 39 | Ccnd2 | 4272 | 0.136 | 0.1406 | No |
| 40 | Ephb2 | 4339 | 0.130 | 0.1398 | No |
| 41 | Lat2 | 4599 | 0.109 | 0.1226 | No |
| 42 | Etv5 | 4624 | 0.107 | 0.1244 | No |
| 43 | Cbl | 4787 | 0.094 | 0.1146 | No |
| 44 | Mmd | 4801 | 0.093 | 0.1168 | No |
| 45 | Eng | 4879 | 0.087 | 0.1136 | No |
| 46 | Zfp277 | 5128 | 0.069 | 0.0959 | No |
| 47 | Cbx8 | 5207 | 0.064 | 0.0919 | No |
| 48 | Cdadc1 | 5255 | 0.060 | 0.0902 | No |
| 49 | Tnfaip3 | 5520 | 0.040 | 0.0702 | No |
| 50 | Map7 | 5676 | 0.031 | 0.0587 | No |
| 51 | Etv1 | 6108 | 0.000 | 0.0238 | No |
| 52 | Cmklr1 | 6430 | -0.005 | -0.0021 | No |
| 53 | Rabgap1l | 6826 | -0.029 | -0.0331 | No |
| 54 | Crot | 6864 | -0.031 | -0.0350 | No |
| 55 | Satb1 | 6869 | -0.031 | -0.0342 | No |
| 56 | Il2rg | 6957 | -0.038 | -0.0399 | No |
| 57 | Atg10 | 7206 | -0.053 | -0.0582 | No |
| 58 | Angptl4 | 7259 | -0.057 | -0.0604 | No |
| 59 | Ptcd2 | 7370 | -0.061 | -0.0672 | No |
| 60 | Ammecr1 | 7493 | -0.068 | -0.0747 | No |
| 61 | Ccser2 | 7523 | -0.070 | -0.0746 | No |
| 62 | Mtmr10 | 7661 | -0.080 | -0.0829 | No |
| 63 | Fcer1g | 7695 | -0.083 | -0.0826 | No |
| 64 | Irf8 | 7924 | -0.100 | -0.0976 | No |
| 65 | Rbm4 | 8190 | -0.117 | -0.1150 | No |
| 66 | Tmem176a | 8318 | -0.127 | -0.1208 | No |
| 67 | Usp12 | 8386 | -0.132 | -0.1216 | No |
| 68 | Tmem176b | 8428 | -0.135 | -0.1202 | No |
| 69 | Dcbld2 | 8633 | -0.149 | -0.1315 | No |
| 70 | Ikzf1 | 9093 | -0.185 | -0.1622 | No |
| 71 | Laptm5 | 9135 | -0.190 | -0.1589 | No |
| 72 | Psmb8 | 9207 | -0.194 | -0.1579 | No |
| 73 | Scg5 | 9265 | -0.199 | -0.1555 | No |
| 74 | Adgrl4 | 9303 | -0.202 | -0.1514 | No |
| 75 | Pecam1 | 9632 | -0.229 | -0.1700 | No |
| 76 | Ly96 | 9683 | -0.234 | -0.1659 | No |
| 77 | Tfpi | 9727 | -0.239 | -0.1610 | No |
| 78 | Zfp639 | 9777 | -0.243 | -0.1564 | No |
| 79 | Map3k1 | 9992 | -0.258 | -0.1647 | No |
| 80 | Gfpt2 | 10046 | -0.264 | -0.1598 | No |
| 81 | Hdac9 | 10136 | -0.266 | -0.1576 | No |
| 82 | Bpgm | 10385 | -0.285 | -0.1677 | No |
| 83 | Pdcd1lg2 | 10738 | -0.323 | -0.1849 | Yes |
| 84 | Nrp1 | 10830 | -0.333 | -0.1806 | Yes |
| 85 | Car2 | 10936 | -0.345 | -0.1770 | Yes |
| 86 | F13a1 | 10962 | -0.349 | -0.1668 | Yes |
| 87 | Fuca1 | 11294 | -0.388 | -0.1801 | Yes |
| 88 | Il10ra | 11301 | -0.390 | -0.1669 | Yes |
| 89 | Dnmbp | 11310 | -0.391 | -0.1538 | Yes |
| 90 | Cxcr4 | 11408 | -0.407 | -0.1475 | Yes |
| 91 | Tspan13 | 11520 | -0.426 | -0.1415 | Yes |
| 92 | Strn | 11696 | -0.458 | -0.1396 | Yes |
| 93 | Kcnn4 | 11789 | -0.473 | -0.1305 | Yes |
| 94 | Prelid3b | 11817 | -0.475 | -0.1160 | Yes |
| 95 | Vwa5a | 11888 | -0.497 | -0.1043 | Yes |
| 96 | Gng11 | 11964 | -0.519 | -0.0922 | Yes |
| 97 | Mycn | 12046 | -0.547 | -0.0796 | Yes |
| 98 | Birc3 | 12071 | -0.557 | -0.0620 | Yes |
| 99 | Csf2ra | 12155 | -0.594 | -0.0479 | Yes |
| 100 | Evi5 | 12168 | -0.599 | -0.0279 | Yes |
| 101 | Id2 | 12291 | -0.680 | -0.0139 | Yes |
| 102 | Sdccag8 | 12343 | -0.742 | 0.0079 | Yes |