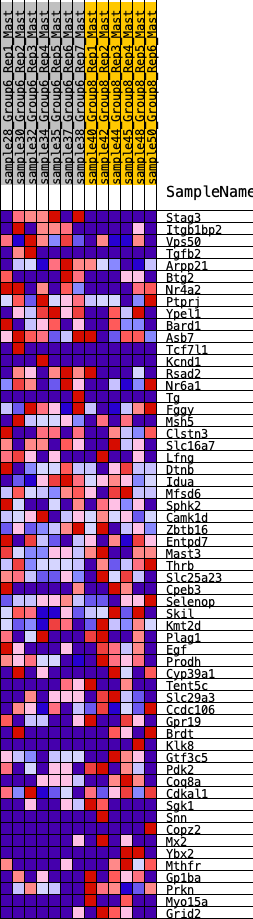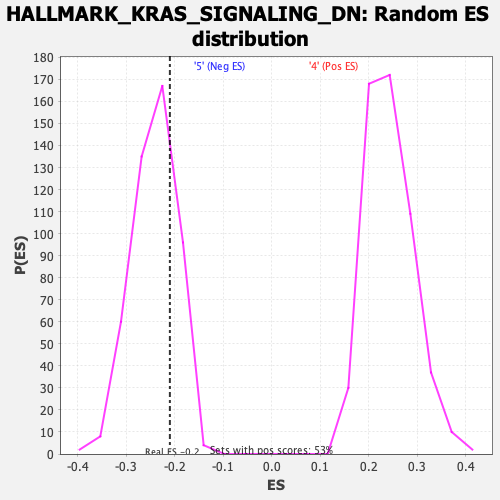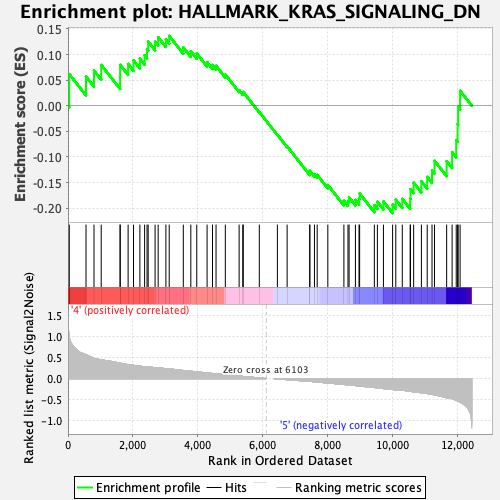
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.20982626 |
| Normalized Enrichment Score (NES) | -0.8686675 |
| Nominal p-value | 0.7288136 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stag3 | 39 | 1.004 | 0.0618 | No |
| 2 | Itgb1bp2 | 555 | 0.568 | 0.0570 | No |
| 3 | Vps50 | 801 | 0.486 | 0.0687 | No |
| 4 | Tgfb2 | 1023 | 0.447 | 0.0797 | No |
| 5 | Arpp21 | 1606 | 0.365 | 0.0563 | No |
| 6 | Btg2 | 1607 | 0.364 | 0.0799 | No |
| 7 | Nr4a2 | 1852 | 0.332 | 0.0817 | No |
| 8 | Ptprj | 2018 | 0.314 | 0.0887 | No |
| 9 | Ypel1 | 2215 | 0.296 | 0.0920 | No |
| 10 | Bard1 | 2360 | 0.281 | 0.0985 | No |
| 11 | Asb7 | 2435 | 0.276 | 0.1104 | No |
| 12 | Tcf7l1 | 2470 | 0.274 | 0.1254 | No |
| 13 | Kcnd1 | 2683 | 0.259 | 0.1250 | No |
| 14 | Rsad2 | 2778 | 0.253 | 0.1338 | No |
| 15 | Nr6a1 | 3016 | 0.234 | 0.1299 | No |
| 16 | Tg | 3118 | 0.228 | 0.1364 | No |
| 17 | Fggy | 3552 | 0.191 | 0.1138 | No |
| 18 | Msh5 | 3784 | 0.174 | 0.1064 | No |
| 19 | Clstn3 | 3967 | 0.160 | 0.1021 | No |
| 20 | Slc16a7 | 4285 | 0.134 | 0.0852 | No |
| 21 | Lfng | 4452 | 0.120 | 0.0795 | No |
| 22 | Dtnb | 4561 | 0.111 | 0.0780 | No |
| 23 | Idua | 4847 | 0.090 | 0.0608 | No |
| 24 | Mfsd6 | 5272 | 0.058 | 0.0303 | No |
| 25 | Sphk2 | 5385 | 0.050 | 0.0246 | No |
| 26 | Camk1d | 5399 | 0.049 | 0.0267 | No |
| 27 | Zbtb16 | 5894 | 0.015 | -0.0123 | No |
| 28 | Entpd7 | 6451 | -0.006 | -0.0568 | No |
| 29 | Mast3 | 6751 | -0.025 | -0.0793 | No |
| 30 | Thrb | 7448 | -0.065 | -0.1313 | No |
| 31 | Slc25a23 | 7450 | -0.065 | -0.1271 | No |
| 32 | Cpeb3 | 7593 | -0.075 | -0.1337 | No |
| 33 | Selenop | 7677 | -0.081 | -0.1351 | No |
| 34 | Skil | 8005 | -0.105 | -0.1548 | No |
| 35 | Kmt2d | 8500 | -0.140 | -0.1856 | No |
| 36 | Plag1 | 8626 | -0.149 | -0.1861 | No |
| 37 | Egf | 8660 | -0.151 | -0.1789 | No |
| 38 | Prodh | 8855 | -0.166 | -0.1839 | No |
| 39 | Cyp39a1 | 8966 | -0.176 | -0.1814 | No |
| 40 | Tent5c | 8985 | -0.177 | -0.1714 | No |
| 41 | Slc29a3 | 9438 | -0.211 | -0.1942 | Yes |
| 42 | Ccdc106 | 9532 | -0.220 | -0.1875 | Yes |
| 43 | Gpr19 | 9718 | -0.238 | -0.1871 | Yes |
| 44 | Brdt | 10001 | -0.259 | -0.1930 | Yes |
| 45 | Klk8 | 10099 | -0.266 | -0.1837 | Yes |
| 46 | Gtf3c5 | 10299 | -0.276 | -0.1819 | Yes |
| 47 | Pdk2 | 10541 | -0.302 | -0.1818 | Yes |
| 48 | Coq8a | 10553 | -0.304 | -0.1630 | Yes |
| 49 | Cdkal1 | 10649 | -0.314 | -0.1503 | Yes |
| 50 | Sgk1 | 10888 | -0.337 | -0.1477 | Yes |
| 51 | Snn | 11067 | -0.357 | -0.1390 | Yes |
| 52 | Copz2 | 11215 | -0.377 | -0.1265 | Yes |
| 53 | Mx2 | 11291 | -0.388 | -0.1075 | Yes |
| 54 | Ybx2 | 11666 | -0.455 | -0.1082 | Yes |
| 55 | Mthfr | 11834 | -0.480 | -0.0906 | Yes |
| 56 | Gp1ba | 11961 | -0.518 | -0.0673 | Yes |
| 57 | Prkn | 12003 | -0.535 | -0.0360 | Yes |
| 58 | Myo15a | 12020 | -0.539 | -0.0024 | Yes |
| 59 | Grid2 | 12080 | -0.562 | 0.0292 | Yes |