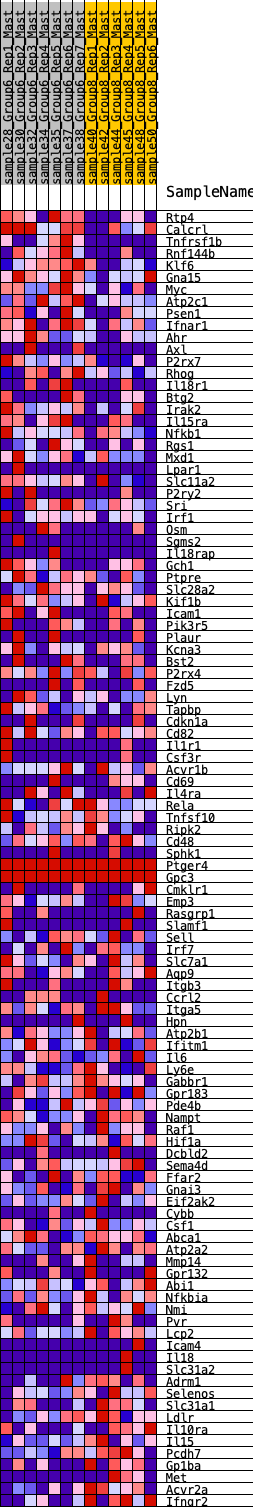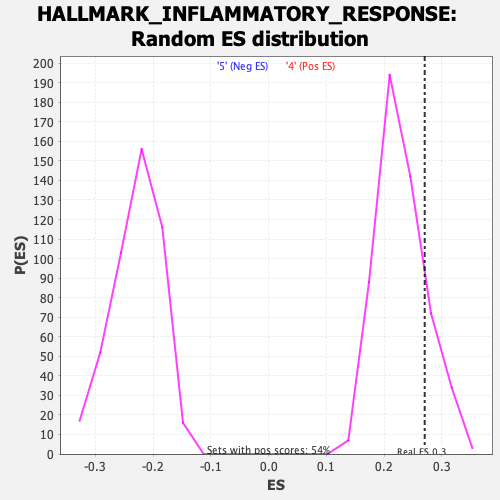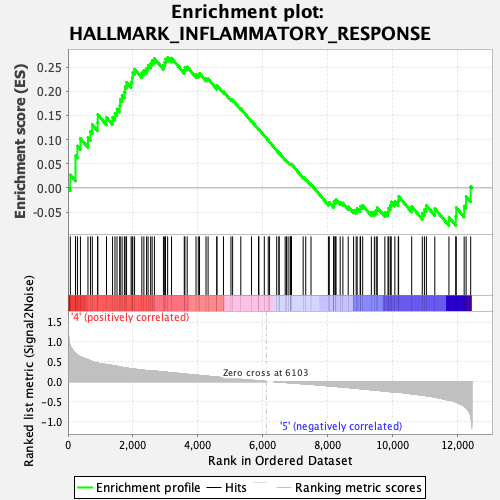
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.27016586 |
| Normalized Enrichment Score (NES) | 1.1795366 |
| Nominal p-value | 0.16481481 |
| FDR q-value | 0.6487579 |
| FWER p-Value | 0.934 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rtp4 | 72 | 0.890 | 0.0271 | Yes |
| 2 | Calcrl | 231 | 0.711 | 0.0406 | Yes |
| 3 | Tnfrsf1b | 232 | 0.709 | 0.0669 | Yes |
| 4 | Rnf144b | 291 | 0.670 | 0.0870 | Yes |
| 5 | Klf6 | 383 | 0.619 | 0.1025 | Yes |
| 6 | Gna15 | 615 | 0.549 | 0.1041 | Yes |
| 7 | Myc | 692 | 0.522 | 0.1173 | Yes |
| 8 | Atp2c1 | 749 | 0.502 | 0.1314 | Yes |
| 9 | Psen1 | 912 | 0.462 | 0.1353 | Yes |
| 10 | Ifnar1 | 918 | 0.461 | 0.1520 | Yes |
| 11 | Ahr | 1186 | 0.426 | 0.1461 | Yes |
| 12 | Axl | 1373 | 0.403 | 0.1460 | Yes |
| 13 | P2rx7 | 1447 | 0.391 | 0.1545 | Yes |
| 14 | Rhog | 1510 | 0.382 | 0.1636 | Yes |
| 15 | Il18r1 | 1594 | 0.367 | 0.1705 | Yes |
| 16 | Btg2 | 1607 | 0.364 | 0.1830 | Yes |
| 17 | Irak2 | 1666 | 0.357 | 0.1915 | Yes |
| 18 | Il15ra | 1737 | 0.346 | 0.1987 | Yes |
| 19 | Nfkb1 | 1759 | 0.344 | 0.2097 | Yes |
| 20 | Rgs1 | 1804 | 0.337 | 0.2187 | Yes |
| 21 | Mxd1 | 1944 | 0.321 | 0.2193 | Yes |
| 22 | Lpar1 | 1977 | 0.318 | 0.2285 | Yes |
| 23 | Slc11a2 | 1996 | 0.317 | 0.2387 | Yes |
| 24 | P2ry2 | 2051 | 0.311 | 0.2459 | Yes |
| 25 | Sri | 2275 | 0.288 | 0.2385 | Yes |
| 26 | Irf1 | 2338 | 0.283 | 0.2439 | Yes |
| 27 | Osm | 2421 | 0.277 | 0.2475 | Yes |
| 28 | Sgms2 | 2467 | 0.274 | 0.2540 | Yes |
| 29 | Il18rap | 2547 | 0.268 | 0.2576 | Yes |
| 30 | Gch1 | 2592 | 0.267 | 0.2639 | Yes |
| 31 | Ptpre | 2662 | 0.260 | 0.2679 | Yes |
| 32 | Slc28a2 | 2940 | 0.241 | 0.2544 | Yes |
| 33 | Kif1b | 2978 | 0.238 | 0.2602 | Yes |
| 34 | Icam1 | 3005 | 0.236 | 0.2669 | Yes |
| 35 | Pik3r5 | 3070 | 0.230 | 0.2702 | Yes |
| 36 | Plaur | 3190 | 0.224 | 0.2688 | No |
| 37 | Kcna3 | 3582 | 0.188 | 0.2441 | No |
| 38 | Bst2 | 3602 | 0.187 | 0.2495 | No |
| 39 | P2rx4 | 3672 | 0.181 | 0.2506 | No |
| 40 | Fzd5 | 3944 | 0.161 | 0.2346 | No |
| 41 | Lyn | 4014 | 0.156 | 0.2348 | No |
| 42 | Tapbp | 4052 | 0.153 | 0.2374 | No |
| 43 | Cdkn1a | 4252 | 0.137 | 0.2264 | No |
| 44 | Cd82 | 4316 | 0.132 | 0.2261 | No |
| 45 | Il1r1 | 4585 | 0.110 | 0.2085 | No |
| 46 | Csf3r | 4587 | 0.110 | 0.2125 | No |
| 47 | Acvr1b | 4790 | 0.094 | 0.1996 | No |
| 48 | Cd69 | 5021 | 0.077 | 0.1838 | No |
| 49 | Il4ra | 5072 | 0.073 | 0.1824 | No |
| 50 | Rela | 5324 | 0.054 | 0.1641 | No |
| 51 | Tnfsf10 | 5653 | 0.032 | 0.1387 | No |
| 52 | Ripk2 | 5869 | 0.016 | 0.1218 | No |
| 53 | Cd48 | 5881 | 0.015 | 0.1215 | No |
| 54 | Sphk1 | 6048 | 0.005 | 0.1082 | No |
| 55 | Ptger4 | 6169 | 0.000 | 0.0985 | No |
| 56 | Gpc3 | 6210 | 0.000 | 0.0953 | No |
| 57 | Cmklr1 | 6430 | -0.005 | 0.0777 | No |
| 58 | Emp3 | 6501 | -0.009 | 0.0723 | No |
| 59 | Rasgrp1 | 6502 | -0.009 | 0.0727 | No |
| 60 | Slamf1 | 6691 | -0.022 | 0.0583 | No |
| 61 | Sell | 6730 | -0.024 | 0.0561 | No |
| 62 | Irf7 | 6781 | -0.026 | 0.0530 | No |
| 63 | Slc7a1 | 6844 | -0.030 | 0.0491 | No |
| 64 | Aqp9 | 6862 | -0.031 | 0.0488 | No |
| 65 | Itgb3 | 6870 | -0.031 | 0.0494 | No |
| 66 | Ccrl2 | 6886 | -0.032 | 0.0494 | No |
| 67 | Itga5 | 7245 | -0.056 | 0.0225 | No |
| 68 | Hpn | 7324 | -0.059 | 0.0183 | No |
| 69 | Atp2b1 | 7487 | -0.068 | 0.0077 | No |
| 70 | Ifitm1 | 8023 | -0.106 | -0.0317 | No |
| 71 | Il6 | 8050 | -0.108 | -0.0298 | No |
| 72 | Ly6e | 8184 | -0.117 | -0.0363 | No |
| 73 | Gabbr1 | 8191 | -0.117 | -0.0324 | No |
| 74 | Gpr183 | 8193 | -0.118 | -0.0281 | No |
| 75 | Pde4b | 8246 | -0.122 | -0.0279 | No |
| 76 | Nampt | 8256 | -0.123 | -0.0240 | No |
| 77 | Raf1 | 8385 | -0.132 | -0.0295 | No |
| 78 | Hif1a | 8469 | -0.138 | -0.0312 | No |
| 79 | Dcbld2 | 8633 | -0.149 | -0.0389 | No |
| 80 | Sema4d | 8799 | -0.162 | -0.0463 | No |
| 81 | Ffar2 | 8878 | -0.168 | -0.0464 | No |
| 82 | Gnai3 | 8908 | -0.170 | -0.0424 | No |
| 83 | Eif2ak2 | 9000 | -0.178 | -0.0432 | No |
| 84 | Cybb | 9013 | -0.179 | -0.0375 | No |
| 85 | Csf1 | 9079 | -0.184 | -0.0360 | No |
| 86 | Abca1 | 9346 | -0.204 | -0.0500 | No |
| 87 | Atp2a2 | 9439 | -0.212 | -0.0496 | No |
| 88 | Mmp14 | 9496 | -0.217 | -0.0461 | No |
| 89 | Gpr132 | 9528 | -0.220 | -0.0405 | No |
| 90 | Abi1 | 9762 | -0.241 | -0.0504 | No |
| 91 | Nfkbia | 9859 | -0.248 | -0.0490 | No |
| 92 | Nmi | 9882 | -0.249 | -0.0416 | No |
| 93 | Pvr | 9931 | -0.253 | -0.0361 | No |
| 94 | Lcp2 | 9958 | -0.256 | -0.0287 | No |
| 95 | Icam4 | 10070 | -0.266 | -0.0279 | No |
| 96 | Il18 | 10175 | -0.267 | -0.0264 | No |
| 97 | Slc31a2 | 10186 | -0.267 | -0.0173 | No |
| 98 | Adrm1 | 10589 | -0.308 | -0.0385 | No |
| 99 | Selenos | 10915 | -0.342 | -0.0522 | No |
| 100 | Slc31a1 | 10981 | -0.349 | -0.0445 | No |
| 101 | Ldlr | 11041 | -0.355 | -0.0362 | No |
| 102 | Il10ra | 11301 | -0.390 | -0.0427 | No |
| 103 | Il15 | 11736 | -0.465 | -0.0607 | No |
| 104 | Pcdh7 | 11946 | -0.511 | -0.0587 | No |
| 105 | Gp1ba | 11961 | -0.518 | -0.0407 | No |
| 106 | Met | 12205 | -0.620 | -0.0374 | No |
| 107 | Acvr2a | 12265 | -0.665 | -0.0176 | No |
| 108 | Ifngr2 | 12406 | -0.857 | 0.0028 | No |