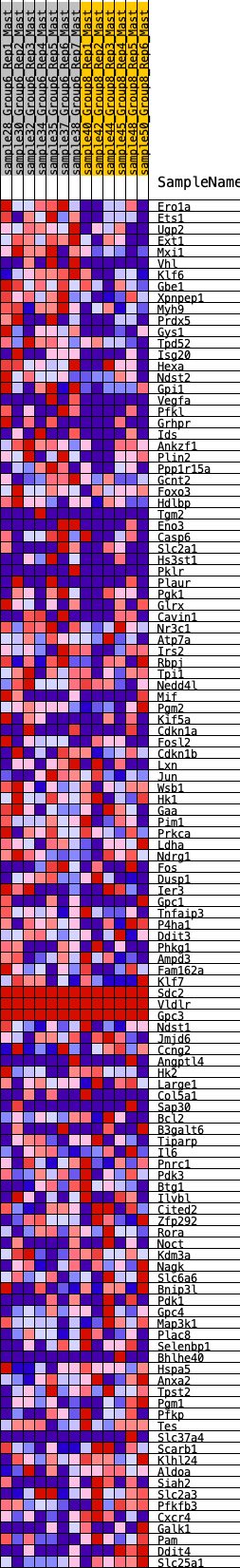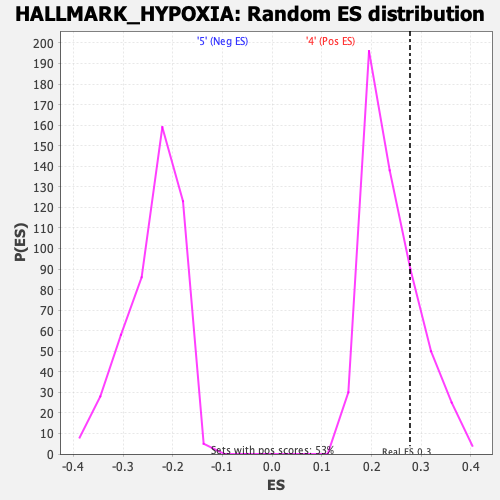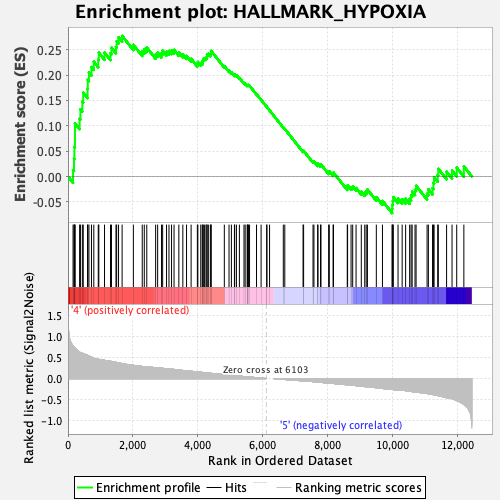
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.27740508 |
| Normalized Enrichment Score (NES) | 1.167559 |
| Nominal p-value | 0.24765478 |
| FDR q-value | 0.5590429 |
| FWER p-Value | 0.944 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ero1a | 156 | 0.773 | 0.0129 | Yes |
| 2 | Ets1 | 185 | 0.747 | 0.0352 | Yes |
| 3 | Ugp2 | 197 | 0.734 | 0.0586 | Yes |
| 4 | Ext1 | 214 | 0.724 | 0.0812 | Yes |
| 5 | Mxi1 | 215 | 0.722 | 0.1050 | Yes |
| 6 | Vhl | 358 | 0.627 | 0.1142 | Yes |
| 7 | Klf6 | 383 | 0.619 | 0.1327 | Yes |
| 8 | Gbe1 | 442 | 0.604 | 0.1479 | Yes |
| 9 | Xpnpep1 | 469 | 0.592 | 0.1653 | Yes |
| 10 | Myh9 | 603 | 0.553 | 0.1728 | Yes |
| 11 | Prdx5 | 609 | 0.552 | 0.1906 | Yes |
| 12 | Gys1 | 645 | 0.536 | 0.2055 | Yes |
| 13 | Tpd52 | 723 | 0.510 | 0.2161 | Yes |
| 14 | Isg20 | 795 | 0.489 | 0.2265 | Yes |
| 15 | Hexa | 932 | 0.458 | 0.2305 | Yes |
| 16 | Ndst2 | 950 | 0.456 | 0.2442 | Yes |
| 17 | Gpi1 | 1125 | 0.435 | 0.2445 | Yes |
| 18 | Vegfa | 1309 | 0.410 | 0.2431 | Yes |
| 19 | Pfkl | 1339 | 0.407 | 0.2542 | Yes |
| 20 | Grhpr | 1478 | 0.386 | 0.2558 | Yes |
| 21 | Ids | 1499 | 0.383 | 0.2668 | Yes |
| 22 | Ankzf1 | 1557 | 0.374 | 0.2745 | Yes |
| 23 | Plin2 | 1667 | 0.356 | 0.2774 | Yes |
| 24 | Ppp1r15a | 2014 | 0.315 | 0.2597 | No |
| 25 | Gcnt2 | 2287 | 0.287 | 0.2471 | No |
| 26 | Foxo3 | 2350 | 0.281 | 0.2514 | No |
| 27 | Hdlbp | 2427 | 0.277 | 0.2544 | No |
| 28 | Tgm2 | 2700 | 0.259 | 0.2408 | No |
| 29 | Eno3 | 2762 | 0.255 | 0.2443 | No |
| 30 | Casp6 | 2880 | 0.247 | 0.2430 | No |
| 31 | Slc2a1 | 2916 | 0.244 | 0.2482 | No |
| 32 | Hs3st1 | 3036 | 0.233 | 0.2462 | No |
| 33 | Pklr | 3111 | 0.228 | 0.2477 | No |
| 34 | Plaur | 3190 | 0.224 | 0.2488 | No |
| 35 | Pgk1 | 3267 | 0.218 | 0.2498 | No |
| 36 | Glrx | 3415 | 0.203 | 0.2446 | No |
| 37 | Cavin1 | 3538 | 0.192 | 0.2410 | No |
| 38 | Nr3c1 | 3652 | 0.183 | 0.2379 | No |
| 39 | Atp7a | 3793 | 0.173 | 0.2323 | No |
| 40 | Irs2 | 3985 | 0.159 | 0.2220 | No |
| 41 | Rbpj | 4001 | 0.158 | 0.2260 | No |
| 42 | Tpi1 | 4090 | 0.149 | 0.2237 | No |
| 43 | Nedd4l | 4135 | 0.146 | 0.2250 | No |
| 44 | Mif | 4152 | 0.145 | 0.2285 | No |
| 45 | Pgm2 | 4168 | 0.143 | 0.2320 | No |
| 46 | Kif5a | 4208 | 0.140 | 0.2335 | No |
| 47 | Cdkn1a | 4252 | 0.137 | 0.2345 | No |
| 48 | Fosl2 | 4282 | 0.135 | 0.2366 | No |
| 49 | Cdkn1b | 4283 | 0.135 | 0.2410 | No |
| 50 | Lxn | 4324 | 0.131 | 0.2421 | No |
| 51 | Jun | 4385 | 0.126 | 0.2414 | No |
| 52 | Wsb1 | 4406 | 0.124 | 0.2438 | No |
| 53 | Hk1 | 4408 | 0.124 | 0.2478 | No |
| 54 | Gaa | 4816 | 0.093 | 0.2179 | No |
| 55 | Pim1 | 4961 | 0.081 | 0.2089 | No |
| 56 | Prkca | 5034 | 0.076 | 0.2055 | No |
| 57 | Ldha | 5127 | 0.069 | 0.2003 | No |
| 58 | Ndrg1 | 5135 | 0.069 | 0.2020 | No |
| 59 | Fos | 5189 | 0.065 | 0.1999 | No |
| 60 | Dusp1 | 5280 | 0.057 | 0.1945 | No |
| 61 | Ier3 | 5422 | 0.048 | 0.1846 | No |
| 62 | Gpc1 | 5458 | 0.045 | 0.1833 | No |
| 63 | Tnfaip3 | 5520 | 0.040 | 0.1796 | No |
| 64 | P4ha1 | 5531 | 0.040 | 0.1801 | No |
| 65 | Ddit3 | 5542 | 0.039 | 0.1806 | No |
| 66 | Phkg1 | 5559 | 0.038 | 0.1805 | No |
| 67 | Ampd3 | 5593 | 0.036 | 0.1791 | No |
| 68 | Fam162a | 5808 | 0.020 | 0.1624 | No |
| 69 | Klf7 | 5949 | 0.011 | 0.1514 | No |
| 70 | Sdc2 | 6123 | 0.000 | 0.1373 | No |
| 71 | Vldlr | 6127 | 0.000 | 0.1371 | No |
| 72 | Gpc3 | 6210 | 0.000 | 0.1304 | No |
| 73 | Ndst1 | 6633 | -0.018 | 0.0968 | No |
| 74 | Jmjd6 | 6679 | -0.021 | 0.0938 | No |
| 75 | Ccng2 | 7242 | -0.055 | 0.0500 | No |
| 76 | Angptl4 | 7259 | -0.057 | 0.0506 | No |
| 77 | Hk2 | 7549 | -0.073 | 0.0295 | No |
| 78 | Large1 | 7577 | -0.075 | 0.0298 | No |
| 79 | Col5a1 | 7692 | -0.083 | 0.0233 | No |
| 80 | Sap30 | 7701 | -0.083 | 0.0254 | No |
| 81 | Bcl2 | 7780 | -0.088 | 0.0220 | No |
| 82 | B3galt6 | 7794 | -0.090 | 0.0239 | No |
| 83 | Tiparp | 8030 | -0.107 | 0.0083 | No |
| 84 | Il6 | 8050 | -0.108 | 0.0103 | No |
| 85 | Pnrc1 | 8167 | -0.115 | 0.0047 | No |
| 86 | Pdk3 | 8180 | -0.117 | 0.0076 | No |
| 87 | Btg1 | 8604 | -0.148 | -0.0218 | No |
| 88 | Ilvbl | 8614 | -0.148 | -0.0177 | No |
| 89 | Cited2 | 8721 | -0.156 | -0.0211 | No |
| 90 | Zfp292 | 8771 | -0.159 | -0.0199 | No |
| 91 | Rora | 8875 | -0.168 | -0.0227 | No |
| 92 | Noct | 9036 | -0.180 | -0.0297 | No |
| 93 | Kdm3a | 9142 | -0.190 | -0.0319 | No |
| 94 | Nagk | 9187 | -0.193 | -0.0292 | No |
| 95 | Slc6a6 | 9225 | -0.195 | -0.0257 | No |
| 96 | Bnip3l | 9499 | -0.218 | -0.0407 | No |
| 97 | Pdk1 | 9688 | -0.234 | -0.0482 | No |
| 98 | Gpc4 | 9986 | -0.258 | -0.0638 | No |
| 99 | Map3k1 | 9992 | -0.258 | -0.0557 | No |
| 100 | Plac8 | 10008 | -0.260 | -0.0483 | No |
| 101 | Selenbp1 | 10021 | -0.261 | -0.0407 | No |
| 102 | Bhlhe40 | 10168 | -0.267 | -0.0437 | No |
| 103 | Hspa5 | 10295 | -0.275 | -0.0448 | No |
| 104 | Anxa2 | 10402 | -0.288 | -0.0439 | No |
| 105 | Tpst2 | 10526 | -0.301 | -0.0440 | No |
| 106 | Pgm1 | 10570 | -0.306 | -0.0374 | No |
| 107 | Pfkp | 10603 | -0.310 | -0.0297 | No |
| 108 | Tes | 10691 | -0.319 | -0.0263 | No |
| 109 | Slc37a4 | 10724 | -0.322 | -0.0182 | No |
| 110 | Scarb1 | 11063 | -0.357 | -0.0339 | No |
| 111 | Klhl24 | 11103 | -0.360 | -0.0252 | No |
| 112 | Aldoa | 11231 | -0.379 | -0.0230 | No |
| 113 | Siah2 | 11258 | -0.383 | -0.0124 | No |
| 114 | Slc2a3 | 11279 | -0.386 | -0.0013 | No |
| 115 | Pfkfb3 | 11391 | -0.403 | 0.0030 | No |
| 116 | Cxcr4 | 11408 | -0.407 | 0.0151 | No |
| 117 | Galk1 | 11664 | -0.455 | 0.0094 | No |
| 118 | Pam | 11832 | -0.480 | 0.0117 | No |
| 119 | Ddit4 | 11976 | -0.524 | 0.0174 | No |
| 120 | Slc25a1 | 12196 | -0.614 | 0.0199 | No |