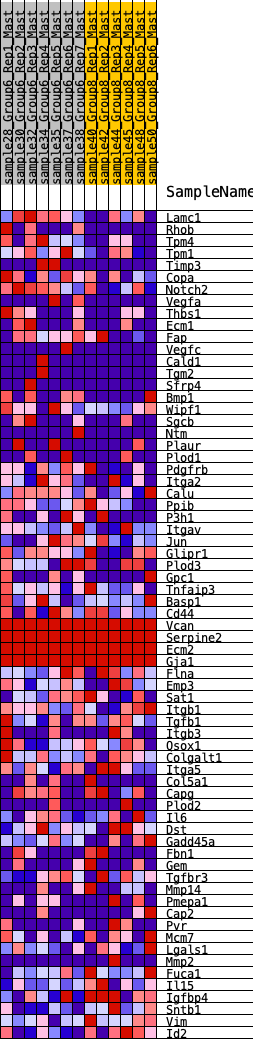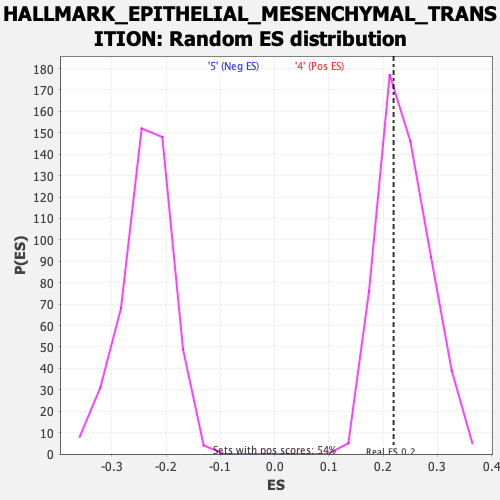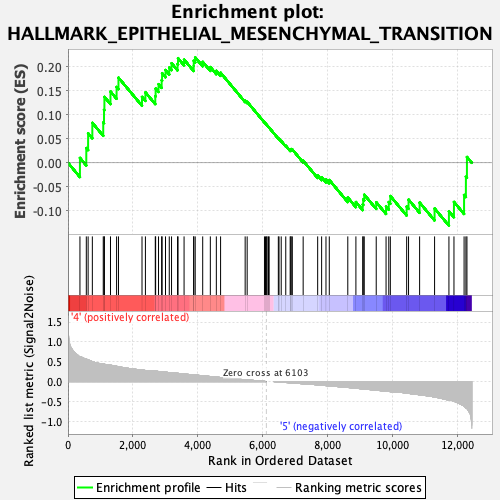
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.21949127 |
| Normalized Enrichment Score (NES) | 0.91949 |
| Nominal p-value | 0.64444447 |
| FDR q-value | 0.8945832 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lamc1 | 368 | 0.623 | 0.0102 | Yes |
| 2 | Rhob | 564 | 0.567 | 0.0308 | Yes |
| 3 | Tpm4 | 620 | 0.548 | 0.0614 | Yes |
| 4 | Tpm1 | 750 | 0.502 | 0.0832 | Yes |
| 5 | Timp3 | 1086 | 0.437 | 0.0842 | Yes |
| 6 | Copa | 1110 | 0.436 | 0.1103 | Yes |
| 7 | Notch2 | 1122 | 0.435 | 0.1373 | Yes |
| 8 | Vegfa | 1309 | 0.410 | 0.1485 | Yes |
| 9 | Thbs1 | 1497 | 0.383 | 0.1580 | Yes |
| 10 | Ecm1 | 1555 | 0.374 | 0.1773 | Yes |
| 11 | Fap | 2280 | 0.288 | 0.1373 | Yes |
| 12 | Vegfc | 2385 | 0.279 | 0.1467 | Yes |
| 13 | Cald1 | 2689 | 0.259 | 0.1388 | Yes |
| 14 | Tgm2 | 2700 | 0.259 | 0.1546 | Yes |
| 15 | Sfrp4 | 2789 | 0.253 | 0.1637 | Yes |
| 16 | Bmp1 | 2885 | 0.247 | 0.1719 | Yes |
| 17 | Wipf1 | 2899 | 0.246 | 0.1866 | Yes |
| 18 | Sgcb | 3004 | 0.236 | 0.1933 | Yes |
| 19 | Ntm | 3120 | 0.228 | 0.1986 | Yes |
| 20 | Plaur | 3190 | 0.224 | 0.2074 | Yes |
| 21 | Plod1 | 3375 | 0.207 | 0.2058 | Yes |
| 22 | Pdgfrb | 3390 | 0.206 | 0.2178 | Yes |
| 23 | Itga2 | 3576 | 0.189 | 0.2150 | Yes |
| 24 | Calu | 3872 | 0.168 | 0.2019 | Yes |
| 25 | Ppib | 3874 | 0.168 | 0.2126 | Yes |
| 26 | P3h1 | 3919 | 0.164 | 0.2195 | Yes |
| 27 | Itgav | 4150 | 0.145 | 0.2102 | No |
| 28 | Jun | 4385 | 0.126 | 0.1994 | No |
| 29 | Glipr1 | 4570 | 0.111 | 0.1916 | No |
| 30 | Plod3 | 4701 | 0.101 | 0.1876 | No |
| 31 | Gpc1 | 5458 | 0.045 | 0.1294 | No |
| 32 | Tnfaip3 | 5520 | 0.040 | 0.1270 | No |
| 33 | Basp1 | 6053 | 0.004 | 0.0843 | No |
| 34 | Cd44 | 6085 | 0.002 | 0.0819 | No |
| 35 | Vcan | 6119 | 0.000 | 0.0792 | No |
| 36 | Serpine2 | 6133 | 0.000 | 0.0782 | No |
| 37 | Ecm2 | 6177 | 0.000 | 0.0747 | No |
| 38 | Gja1 | 6197 | 0.000 | 0.0731 | No |
| 39 | Flna | 6480 | -0.008 | 0.0509 | No |
| 40 | Emp3 | 6501 | -0.009 | 0.0498 | No |
| 41 | Sat1 | 6569 | -0.014 | 0.0453 | No |
| 42 | Itgb1 | 6709 | -0.023 | 0.0356 | No |
| 43 | Tgfb1 | 6845 | -0.030 | 0.0266 | No |
| 44 | Itgb3 | 6870 | -0.031 | 0.0267 | No |
| 45 | Qsox1 | 6871 | -0.031 | 0.0287 | No |
| 46 | Colgalt1 | 6911 | -0.034 | 0.0277 | No |
| 47 | Itga5 | 7245 | -0.056 | 0.0044 | No |
| 48 | Col5a1 | 7692 | -0.083 | -0.0264 | No |
| 49 | Capg | 7814 | -0.091 | -0.0303 | No |
| 50 | Plod2 | 7948 | -0.101 | -0.0346 | No |
| 51 | Il6 | 8050 | -0.108 | -0.0358 | No |
| 52 | Dst | 8620 | -0.149 | -0.0723 | No |
| 53 | Gadd45a | 8870 | -0.167 | -0.0817 | No |
| 54 | Fbn1 | 9080 | -0.184 | -0.0868 | No |
| 55 | Gem | 9096 | -0.186 | -0.0761 | No |
| 56 | Tgfbr3 | 9127 | -0.189 | -0.0664 | No |
| 57 | Mmp14 | 9496 | -0.217 | -0.0822 | No |
| 58 | Pmepa1 | 9799 | -0.245 | -0.0909 | No |
| 59 | Cap2 | 9883 | -0.249 | -0.0816 | No |
| 60 | Pvr | 9931 | -0.253 | -0.0692 | No |
| 61 | Mcm7 | 10434 | -0.291 | -0.0911 | No |
| 62 | Lgals1 | 10492 | -0.297 | -0.0767 | No |
| 63 | Mmp2 | 10833 | -0.333 | -0.0828 | No |
| 64 | Fuca1 | 11294 | -0.388 | -0.0951 | No |
| 65 | Il15 | 11736 | -0.465 | -0.1010 | No |
| 66 | Igfbp4 | 11891 | -0.498 | -0.0815 | No |
| 67 | Sntb1 | 12203 | -0.619 | -0.0669 | No |
| 68 | Vim | 12259 | -0.662 | -0.0290 | No |
| 69 | Id2 | 12291 | -0.680 | 0.0121 | No |