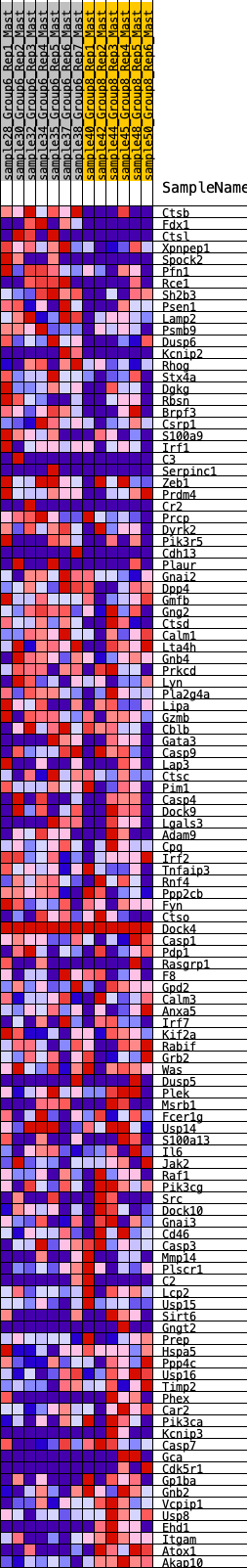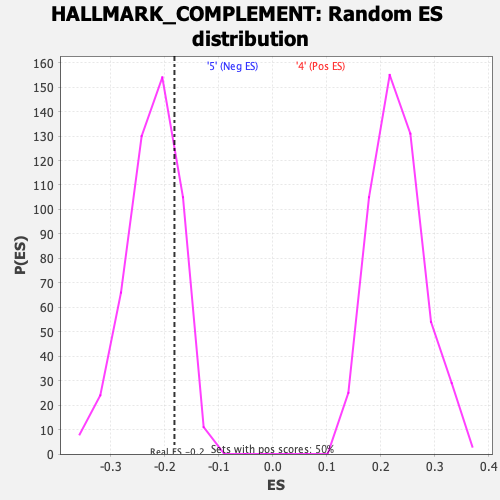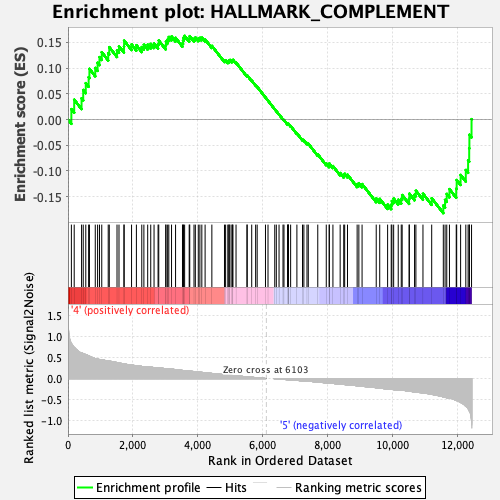
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.18195024 |
| Normalized Enrichment Score (NES) | -0.8161957 |
| Nominal p-value | 0.8072289 |
| FDR q-value | 0.9002218 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ctsb | 104 | 0.835 | 0.0202 | No |
| 2 | Fdx1 | 191 | 0.739 | 0.0385 | No |
| 3 | Ctsl | 418 | 0.613 | 0.0412 | No |
| 4 | Xpnpep1 | 469 | 0.592 | 0.0574 | No |
| 5 | Spock2 | 548 | 0.571 | 0.0706 | No |
| 6 | Pfn1 | 636 | 0.540 | 0.0821 | No |
| 7 | Rce1 | 660 | 0.531 | 0.0984 | No |
| 8 | Sh2b3 | 840 | 0.476 | 0.1002 | No |
| 9 | Psen1 | 912 | 0.462 | 0.1103 | No |
| 10 | Lamp2 | 970 | 0.454 | 0.1212 | No |
| 11 | Psmb9 | 1040 | 0.444 | 0.1308 | No |
| 12 | Dusp6 | 1239 | 0.417 | 0.1290 | No |
| 13 | Kcnip2 | 1272 | 0.413 | 0.1406 | No |
| 14 | Rhog | 1510 | 0.382 | 0.1344 | No |
| 15 | Stx4a | 1571 | 0.371 | 0.1423 | No |
| 16 | Dgkg | 1724 | 0.348 | 0.1419 | No |
| 17 | Rbsn | 1729 | 0.347 | 0.1534 | No |
| 18 | Brpf3 | 1958 | 0.320 | 0.1459 | No |
| 19 | Csrp1 | 2107 | 0.305 | 0.1443 | No |
| 20 | S100a9 | 2273 | 0.289 | 0.1408 | No |
| 21 | Irf1 | 2338 | 0.283 | 0.1453 | No |
| 22 | C3 | 2455 | 0.274 | 0.1453 | No |
| 23 | Serpinc1 | 2549 | 0.268 | 0.1470 | No |
| 24 | Zeb1 | 2651 | 0.261 | 0.1477 | No |
| 25 | Prdm4 | 2775 | 0.254 | 0.1464 | No |
| 26 | Cr2 | 2796 | 0.253 | 0.1535 | No |
| 27 | Prcp | 3011 | 0.235 | 0.1442 | No |
| 28 | Dyrk2 | 3019 | 0.234 | 0.1516 | No |
| 29 | Pik3r5 | 3070 | 0.230 | 0.1554 | No |
| 30 | Cdh13 | 3106 | 0.228 | 0.1604 | No |
| 31 | Plaur | 3190 | 0.224 | 0.1614 | No |
| 32 | Gnai2 | 3314 | 0.213 | 0.1587 | No |
| 33 | Dpp4 | 3527 | 0.193 | 0.1481 | No |
| 34 | Gmfb | 3545 | 0.192 | 0.1533 | No |
| 35 | Gng2 | 3557 | 0.191 | 0.1589 | No |
| 36 | Ctsd | 3593 | 0.188 | 0.1625 | No |
| 37 | Calm1 | 3734 | 0.177 | 0.1572 | No |
| 38 | Lta4h | 3751 | 0.176 | 0.1620 | No |
| 39 | Gnb4 | 3882 | 0.166 | 0.1571 | No |
| 40 | Prkcd | 3923 | 0.163 | 0.1594 | No |
| 41 | Lyn | 4014 | 0.156 | 0.1575 | No |
| 42 | Pla2g4a | 4063 | 0.151 | 0.1588 | No |
| 43 | Lipa | 4123 | 0.147 | 0.1590 | No |
| 44 | Gzmb | 4224 | 0.139 | 0.1557 | No |
| 45 | Cblb | 4432 | 0.122 | 0.1431 | No |
| 46 | Gata3 | 4821 | 0.092 | 0.1147 | No |
| 47 | Casp9 | 4856 | 0.090 | 0.1150 | No |
| 48 | Lap3 | 4924 | 0.084 | 0.1125 | No |
| 49 | Ctsc | 4958 | 0.081 | 0.1126 | No |
| 50 | Pim1 | 4961 | 0.081 | 0.1152 | No |
| 51 | Casp4 | 4989 | 0.079 | 0.1157 | No |
| 52 | Dock9 | 5050 | 0.075 | 0.1134 | No |
| 53 | Lgals3 | 5064 | 0.074 | 0.1149 | No |
| 54 | Adam9 | 5082 | 0.072 | 0.1160 | No |
| 55 | Cpq | 5177 | 0.066 | 0.1106 | No |
| 56 | Irf2 | 5514 | 0.041 | 0.0848 | No |
| 57 | Tnfaip3 | 5520 | 0.040 | 0.0857 | No |
| 58 | Rnf4 | 5660 | 0.032 | 0.0755 | No |
| 59 | Ppp2cb | 5781 | 0.022 | 0.0666 | No |
| 60 | Fyn | 5828 | 0.019 | 0.0635 | No |
| 61 | Ctso | 6084 | 0.002 | 0.0429 | No |
| 62 | Dock4 | 6162 | 0.000 | 0.0366 | No |
| 63 | Casp1 | 6370 | -0.002 | 0.0199 | No |
| 64 | Pdp1 | 6418 | -0.004 | 0.0162 | No |
| 65 | Rasgrp1 | 6502 | -0.009 | 0.0098 | No |
| 66 | F8 | 6624 | -0.017 | 0.0006 | No |
| 67 | Gpd2 | 6654 | -0.019 | -0.0011 | No |
| 68 | Calm3 | 6764 | -0.025 | -0.0091 | No |
| 69 | Anxa5 | 6780 | -0.026 | -0.0094 | No |
| 70 | Irf7 | 6781 | -0.026 | -0.0085 | No |
| 71 | Kif2a | 6792 | -0.027 | -0.0084 | No |
| 72 | Rabif | 6859 | -0.031 | -0.0127 | No |
| 73 | Grb2 | 7052 | -0.043 | -0.0268 | No |
| 74 | Was | 7225 | -0.054 | -0.0389 | No |
| 75 | Dusp5 | 7267 | -0.057 | -0.0403 | No |
| 76 | Plek | 7363 | -0.060 | -0.0459 | No |
| 77 | Msrb1 | 7403 | -0.063 | -0.0469 | No |
| 78 | Fcer1g | 7695 | -0.083 | -0.0677 | No |
| 79 | Usp14 | 7957 | -0.101 | -0.0854 | No |
| 80 | S100a13 | 8048 | -0.108 | -0.0890 | No |
| 81 | Il6 | 8050 | -0.108 | -0.0854 | No |
| 82 | Jak2 | 8162 | -0.115 | -0.0905 | No |
| 83 | Raf1 | 8385 | -0.132 | -0.1040 | No |
| 84 | Pik3cg | 8495 | -0.139 | -0.1081 | No |
| 85 | Src | 8524 | -0.142 | -0.1055 | No |
| 86 | Dock10 | 8611 | -0.148 | -0.1074 | No |
| 87 | Gnai3 | 8908 | -0.170 | -0.1255 | No |
| 88 | Cd46 | 8961 | -0.175 | -0.1237 | No |
| 89 | Casp3 | 9060 | -0.182 | -0.1255 | No |
| 90 | Mmp14 | 9496 | -0.217 | -0.1533 | No |
| 91 | Plscr1 | 9604 | -0.226 | -0.1542 | No |
| 92 | C2 | 9847 | -0.248 | -0.1654 | No |
| 93 | Lcp2 | 9958 | -0.256 | -0.1655 | No |
| 94 | Usp15 | 9979 | -0.257 | -0.1583 | No |
| 95 | Sirt6 | 10033 | -0.262 | -0.1536 | No |
| 96 | Gngt2 | 10174 | -0.267 | -0.1558 | No |
| 97 | Prep | 10267 | -0.272 | -0.1540 | No |
| 98 | Hspa5 | 10295 | -0.275 | -0.1467 | No |
| 99 | Ppp4c | 10509 | -0.299 | -0.1538 | No |
| 100 | Usp16 | 10517 | -0.299 | -0.1441 | No |
| 101 | Timp2 | 10677 | -0.317 | -0.1461 | No |
| 102 | Phex | 10718 | -0.322 | -0.1383 | No |
| 103 | Car2 | 10936 | -0.345 | -0.1441 | No |
| 104 | Pik3ca | 11205 | -0.376 | -0.1530 | No |
| 105 | Kcnip3 | 11563 | -0.434 | -0.1671 | Yes |
| 106 | Casp7 | 11617 | -0.448 | -0.1560 | Yes |
| 107 | Gca | 11669 | -0.455 | -0.1446 | Yes |
| 108 | Cdk5r1 | 11752 | -0.466 | -0.1353 | Yes |
| 109 | Gp1ba | 11961 | -0.518 | -0.1344 | Yes |
| 110 | Gnb2 | 11975 | -0.523 | -0.1175 | Yes |
| 111 | Vcpip1 | 12096 | -0.569 | -0.1078 | Yes |
| 112 | Usp8 | 12256 | -0.659 | -0.0981 | Yes |
| 113 | Ehd1 | 12327 | -0.713 | -0.0793 | Yes |
| 114 | Itgam | 12361 | -0.766 | -0.0558 | Yes |
| 115 | Atox1 | 12367 | -0.775 | -0.0296 | Yes |
| 116 | Akap10 | 12433 | -1.037 | 0.0006 | Yes |