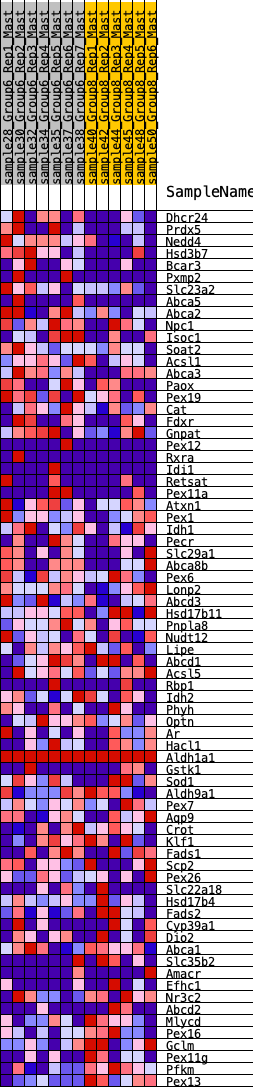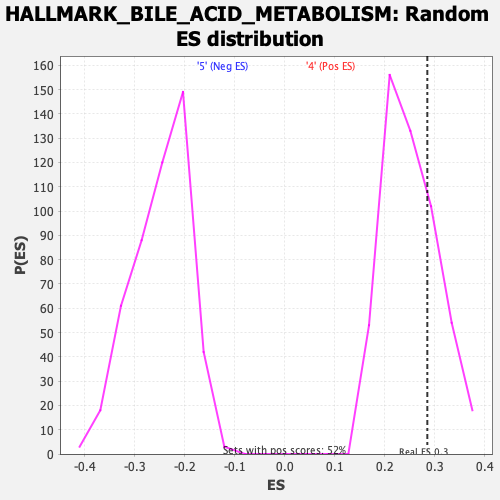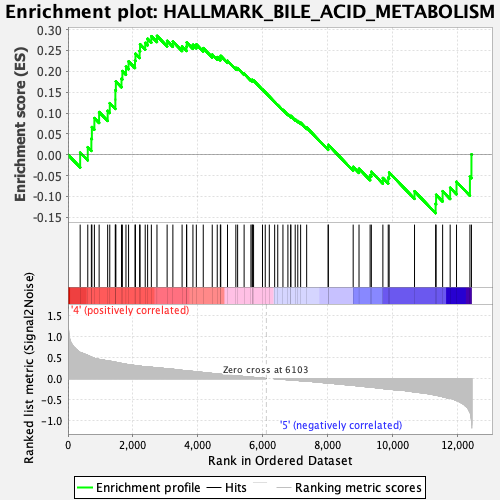
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.2849854 |
| Normalized Enrichment Score (NES) | 1.1319219 |
| Nominal p-value | 0.2771318 |
| FDR q-value | 0.6236491 |
| FWER p-Value | 0.972 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 375 | 0.620 | 0.0053 | Yes |
| 2 | Prdx5 | 609 | 0.552 | 0.0181 | Yes |
| 3 | Nedd4 | 720 | 0.511 | 0.0385 | Yes |
| 4 | Hsd3b7 | 736 | 0.506 | 0.0663 | Yes |
| 5 | Bcar3 | 813 | 0.483 | 0.0879 | Yes |
| 6 | Pxmp2 | 959 | 0.456 | 0.1023 | Yes |
| 7 | Slc23a2 | 1222 | 0.419 | 0.1052 | Yes |
| 8 | Abca5 | 1287 | 0.412 | 0.1236 | Yes |
| 9 | Abca2 | 1459 | 0.390 | 0.1322 | Yes |
| 10 | Npc1 | 1460 | 0.390 | 0.1546 | Yes |
| 11 | Isoc1 | 1475 | 0.387 | 0.1756 | Yes |
| 12 | Soat2 | 1648 | 0.359 | 0.1823 | Yes |
| 13 | Acsl1 | 1675 | 0.355 | 0.2006 | Yes |
| 14 | Abca3 | 1786 | 0.340 | 0.2112 | Yes |
| 15 | Paox | 1866 | 0.330 | 0.2238 | Yes |
| 16 | Pex19 | 2064 | 0.310 | 0.2256 | Yes |
| 17 | Cat | 2079 | 0.308 | 0.2422 | Yes |
| 18 | Fdxr | 2207 | 0.297 | 0.2489 | Yes |
| 19 | Gnpat | 2220 | 0.295 | 0.2649 | Yes |
| 20 | Pex12 | 2379 | 0.279 | 0.2681 | Yes |
| 21 | Rxra | 2453 | 0.274 | 0.2779 | Yes |
| 22 | Idi1 | 2568 | 0.268 | 0.2841 | Yes |
| 23 | Retsat | 2741 | 0.258 | 0.2850 | Yes |
| 24 | Pex11a | 3054 | 0.231 | 0.2730 | No |
| 25 | Atxn1 | 3231 | 0.221 | 0.2714 | No |
| 26 | Pex1 | 3515 | 0.194 | 0.2597 | No |
| 27 | Idh1 | 3651 | 0.183 | 0.2593 | No |
| 28 | Pecr | 3658 | 0.183 | 0.2693 | No |
| 29 | Slc29a1 | 3849 | 0.169 | 0.2637 | No |
| 30 | Abca8b | 3954 | 0.161 | 0.2645 | No |
| 31 | Pex6 | 4167 | 0.143 | 0.2556 | No |
| 32 | Lonp2 | 4444 | 0.121 | 0.2402 | No |
| 33 | Abcd3 | 4598 | 0.109 | 0.2340 | No |
| 34 | Hsd17b11 | 4694 | 0.101 | 0.2322 | No |
| 35 | Pnpla8 | 4706 | 0.100 | 0.2370 | No |
| 36 | Nudt12 | 4914 | 0.085 | 0.2252 | No |
| 37 | Lipe | 5171 | 0.067 | 0.2083 | No |
| 38 | Abcd1 | 5226 | 0.063 | 0.2076 | No |
| 39 | Acsl5 | 5426 | 0.048 | 0.1942 | No |
| 40 | Rbp1 | 5636 | 0.034 | 0.1793 | No |
| 41 | Idh2 | 5677 | 0.031 | 0.1778 | No |
| 42 | Phyh | 5678 | 0.031 | 0.1796 | No |
| 43 | Optn | 5717 | 0.028 | 0.1781 | No |
| 44 | Ar | 5992 | 0.010 | 0.1565 | No |
| 45 | Hacl1 | 6077 | 0.002 | 0.1498 | No |
| 46 | Aldh1a1 | 6202 | 0.000 | 0.1398 | No |
| 47 | Gstk1 | 6367 | -0.002 | 0.1266 | No |
| 48 | Sod1 | 6462 | -0.007 | 0.1194 | No |
| 49 | Aldh9a1 | 6622 | -0.017 | 0.1075 | No |
| 50 | Pex7 | 6775 | -0.026 | 0.0967 | No |
| 51 | Aqp9 | 6862 | -0.031 | 0.0915 | No |
| 52 | Crot | 6864 | -0.031 | 0.0932 | No |
| 53 | Klf1 | 7000 | -0.040 | 0.0846 | No |
| 54 | Fads1 | 7079 | -0.045 | 0.0809 | No |
| 55 | Scp2 | 7167 | -0.050 | 0.0767 | No |
| 56 | Pex26 | 7353 | -0.060 | 0.0652 | No |
| 57 | Slc22a18 | 8016 | -0.106 | 0.0178 | No |
| 58 | Hsd17b4 | 8019 | -0.106 | 0.0237 | No |
| 59 | Fads2 | 8786 | -0.160 | -0.0291 | No |
| 60 | Cyp39a1 | 8966 | -0.176 | -0.0335 | No |
| 61 | Dio2 | 9308 | -0.202 | -0.0494 | No |
| 62 | Abca1 | 9346 | -0.204 | -0.0407 | No |
| 63 | Slc35b2 | 9701 | -0.236 | -0.0558 | No |
| 64 | Amacr | 9866 | -0.249 | -0.0548 | No |
| 65 | Efhc1 | 9894 | -0.250 | -0.0426 | No |
| 66 | Nr3c2 | 10676 | -0.317 | -0.0875 | No |
| 67 | Abcd2 | 11326 | -0.392 | -0.1175 | No |
| 68 | Mlycd | 11341 | -0.394 | -0.0960 | No |
| 69 | Pex16 | 11542 | -0.431 | -0.0874 | No |
| 70 | Gclm | 11774 | -0.471 | -0.0791 | No |
| 71 | Pex11g | 11970 | -0.521 | -0.0649 | No |
| 72 | Pfkm | 12383 | -0.798 | -0.0524 | No |
| 73 | Pex13 | 12430 | -0.994 | 0.0009 | No |