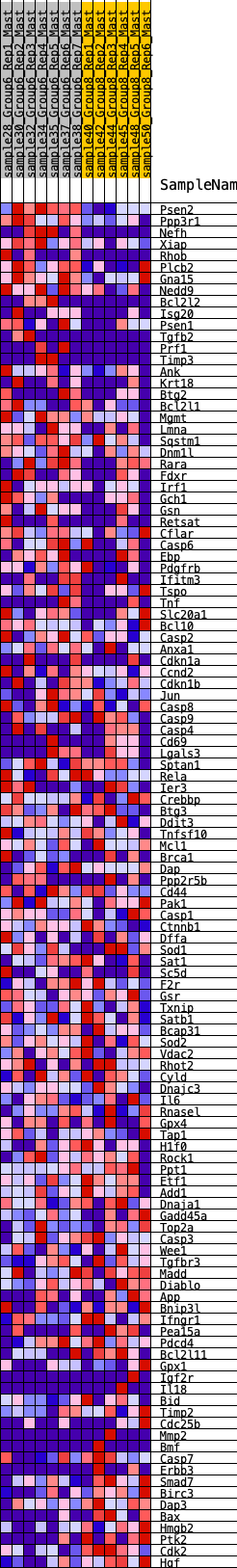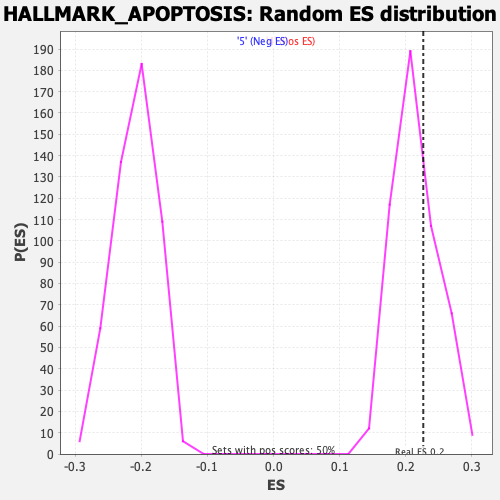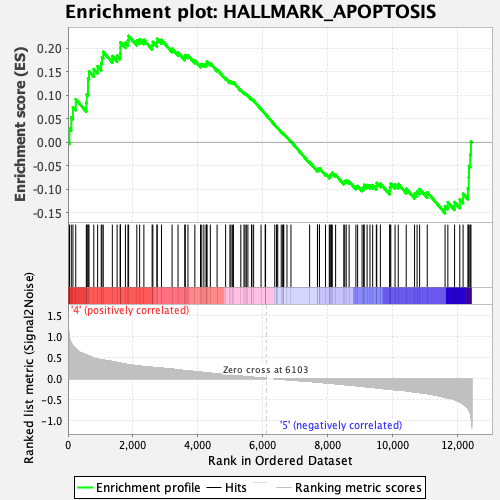
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group6_versus_Group8.Mast_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Mast_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.22658329 |
| Normalized Enrichment Score (NES) | 1.0549649 |
| Nominal p-value | 0.334 |
| FDR q-value | 0.7224442 |
| FWER p-Value | 0.987 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Psen2 | 46 | 0.977 | 0.0291 | Yes |
| 2 | Ppp3r1 | 101 | 0.837 | 0.0528 | Yes |
| 3 | Nefh | 152 | 0.776 | 0.0747 | Yes |
| 4 | Xiap | 239 | 0.707 | 0.0915 | Yes |
| 5 | Rhob | 564 | 0.567 | 0.0842 | Yes |
| 6 | Plcb2 | 578 | 0.560 | 0.1020 | Yes |
| 7 | Gna15 | 615 | 0.549 | 0.1175 | Yes |
| 8 | Nedd9 | 617 | 0.548 | 0.1358 | Yes |
| 9 | Bcl2l2 | 647 | 0.536 | 0.1514 | Yes |
| 10 | Isg20 | 795 | 0.489 | 0.1559 | Yes |
| 11 | Psen1 | 912 | 0.462 | 0.1620 | Yes |
| 12 | Tgfb2 | 1023 | 0.447 | 0.1681 | Yes |
| 13 | Prf1 | 1047 | 0.443 | 0.1810 | Yes |
| 14 | Timp3 | 1086 | 0.437 | 0.1926 | Yes |
| 15 | Ank | 1366 | 0.404 | 0.1836 | Yes |
| 16 | Krt18 | 1513 | 0.381 | 0.1845 | Yes |
| 17 | Btg2 | 1607 | 0.364 | 0.1892 | Yes |
| 18 | Bcl2l1 | 1615 | 0.363 | 0.2008 | Yes |
| 19 | Mgmt | 1617 | 0.363 | 0.2129 | Yes |
| 20 | Lmna | 1769 | 0.343 | 0.2122 | Yes |
| 21 | Sqstm1 | 1847 | 0.333 | 0.2171 | Yes |
| 22 | Dnm1l | 1868 | 0.330 | 0.2266 | Yes |
| 23 | Rara | 2120 | 0.303 | 0.2164 | No |
| 24 | Fdxr | 2207 | 0.297 | 0.2194 | No |
| 25 | Irf1 | 2338 | 0.283 | 0.2183 | No |
| 26 | Gch1 | 2592 | 0.267 | 0.2068 | No |
| 27 | Gsn | 2615 | 0.265 | 0.2139 | No |
| 28 | Retsat | 2741 | 0.258 | 0.2124 | No |
| 29 | Cflar | 2747 | 0.257 | 0.2206 | No |
| 30 | Casp6 | 2880 | 0.247 | 0.2182 | No |
| 31 | Ebp | 3207 | 0.222 | 0.1992 | No |
| 32 | Pdgfrb | 3390 | 0.206 | 0.1913 | No |
| 33 | Ifitm3 | 3597 | 0.187 | 0.1809 | No |
| 34 | Tspo | 3614 | 0.186 | 0.1859 | No |
| 35 | Tnf | 3696 | 0.180 | 0.1853 | No |
| 36 | Slc20a1 | 3908 | 0.164 | 0.1737 | No |
| 37 | Bcl10 | 4082 | 0.150 | 0.1647 | No |
| 38 | Casp2 | 4121 | 0.147 | 0.1665 | No |
| 39 | Anxa1 | 4186 | 0.141 | 0.1661 | No |
| 40 | Cdkn1a | 4252 | 0.137 | 0.1654 | No |
| 41 | Ccnd2 | 4272 | 0.136 | 0.1684 | No |
| 42 | Cdkn1b | 4283 | 0.135 | 0.1721 | No |
| 43 | Jun | 4385 | 0.126 | 0.1681 | No |
| 44 | Casp8 | 4593 | 0.110 | 0.1550 | No |
| 45 | Casp9 | 4856 | 0.090 | 0.1368 | No |
| 46 | Casp4 | 4989 | 0.079 | 0.1287 | No |
| 47 | Cd69 | 5021 | 0.077 | 0.1288 | No |
| 48 | Lgals3 | 5064 | 0.074 | 0.1279 | No |
| 49 | Sptan1 | 5097 | 0.071 | 0.1276 | No |
| 50 | Rela | 5324 | 0.054 | 0.1111 | No |
| 51 | Ier3 | 5422 | 0.048 | 0.1049 | No |
| 52 | Crebbp | 5454 | 0.045 | 0.1039 | No |
| 53 | Btg3 | 5494 | 0.042 | 0.1021 | No |
| 54 | Ddit3 | 5542 | 0.039 | 0.0996 | No |
| 55 | Tnfsf10 | 5653 | 0.032 | 0.0917 | No |
| 56 | Mcl1 | 5666 | 0.031 | 0.0918 | No |
| 57 | Brca1 | 5719 | 0.027 | 0.0885 | No |
| 58 | Dap | 5948 | 0.011 | 0.0704 | No |
| 59 | Ppp2r5b | 6075 | 0.002 | 0.0602 | No |
| 60 | Cd44 | 6085 | 0.002 | 0.0596 | No |
| 61 | Pak1 | 6087 | 0.001 | 0.0595 | No |
| 62 | Casp1 | 6370 | -0.002 | 0.0367 | No |
| 63 | Ctnnb1 | 6427 | -0.005 | 0.0323 | No |
| 64 | Dffa | 6432 | -0.005 | 0.0322 | No |
| 65 | Sod1 | 6462 | -0.007 | 0.0300 | No |
| 66 | Sat1 | 6569 | -0.014 | 0.0219 | No |
| 67 | Sc5d | 6613 | -0.016 | 0.0190 | No |
| 68 | F2r | 6642 | -0.019 | 0.0173 | No |
| 69 | Gsr | 6646 | -0.019 | 0.0177 | No |
| 70 | Txnip | 6746 | -0.025 | 0.0105 | No |
| 71 | Satb1 | 6869 | -0.031 | 0.0017 | No |
| 72 | Bcap31 | 7444 | -0.065 | -0.0427 | No |
| 73 | Sod2 | 7685 | -0.082 | -0.0594 | No |
| 74 | Vdac2 | 7687 | -0.082 | -0.0568 | No |
| 75 | Rhot2 | 7746 | -0.086 | -0.0586 | No |
| 76 | Cyld | 7748 | -0.086 | -0.0558 | No |
| 77 | Dnajc3 | 7933 | -0.100 | -0.0673 | No |
| 78 | Il6 | 8050 | -0.108 | -0.0731 | No |
| 79 | Rnasel | 8070 | -0.109 | -0.0710 | No |
| 80 | Gpx4 | 8096 | -0.111 | -0.0693 | No |
| 81 | Tap1 | 8120 | -0.112 | -0.0674 | No |
| 82 | H1f0 | 8141 | -0.114 | -0.0652 | No |
| 83 | Rock1 | 8245 | -0.122 | -0.0695 | No |
| 84 | Ppt1 | 8497 | -0.140 | -0.0852 | No |
| 85 | Etf1 | 8527 | -0.142 | -0.0828 | No |
| 86 | Add1 | 8572 | -0.146 | -0.0814 | No |
| 87 | Dnaja1 | 8661 | -0.151 | -0.0835 | No |
| 88 | Gadd45a | 8870 | -0.167 | -0.0948 | No |
| 89 | Top2a | 8923 | -0.172 | -0.0932 | No |
| 90 | Casp3 | 9060 | -0.182 | -0.0981 | No |
| 91 | Wee1 | 9109 | -0.187 | -0.0958 | No |
| 92 | Tgfbr3 | 9127 | -0.189 | -0.0908 | No |
| 93 | Madd | 9215 | -0.195 | -0.0913 | No |
| 94 | Diablo | 9304 | -0.202 | -0.0917 | No |
| 95 | App | 9387 | -0.207 | -0.0914 | No |
| 96 | Bnip3l | 9499 | -0.218 | -0.0931 | No |
| 97 | Ifngr1 | 9513 | -0.219 | -0.0868 | No |
| 98 | Pea15a | 9624 | -0.228 | -0.0881 | No |
| 99 | Pdcd4 | 9909 | -0.251 | -0.1027 | No |
| 100 | Bcl2l11 | 9921 | -0.252 | -0.0951 | No |
| 101 | Gpx1 | 9945 | -0.254 | -0.0885 | No |
| 102 | Igf2r | 10076 | -0.266 | -0.0901 | No |
| 103 | Il18 | 10175 | -0.267 | -0.0891 | No |
| 104 | Bid | 10420 | -0.289 | -0.0992 | No |
| 105 | Timp2 | 10677 | -0.317 | -0.1093 | No |
| 106 | Cdc25b | 10755 | -0.324 | -0.1047 | No |
| 107 | Mmp2 | 10833 | -0.333 | -0.0997 | No |
| 108 | Bmf | 11068 | -0.357 | -0.1067 | No |
| 109 | Casp7 | 11617 | -0.448 | -0.1362 | No |
| 110 | Erbb3 | 11703 | -0.460 | -0.1276 | No |
| 111 | Smad7 | 11910 | -0.502 | -0.1275 | No |
| 112 | Birc3 | 12071 | -0.557 | -0.1218 | No |
| 113 | Dap3 | 12172 | -0.600 | -0.1098 | No |
| 114 | Bax | 12321 | -0.709 | -0.0980 | No |
| 115 | Hmgb2 | 12347 | -0.744 | -0.0750 | No |
| 116 | Ptk2 | 12353 | -0.751 | -0.0503 | No |
| 117 | Cdk2 | 12393 | -0.813 | -0.0261 | No |
| 118 | Hgf | 12415 | -0.892 | 0.0021 | No |