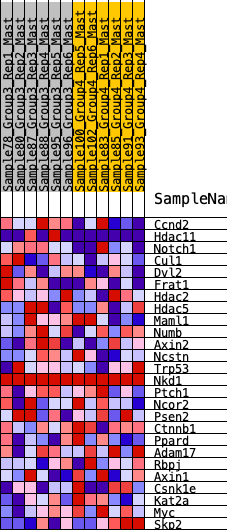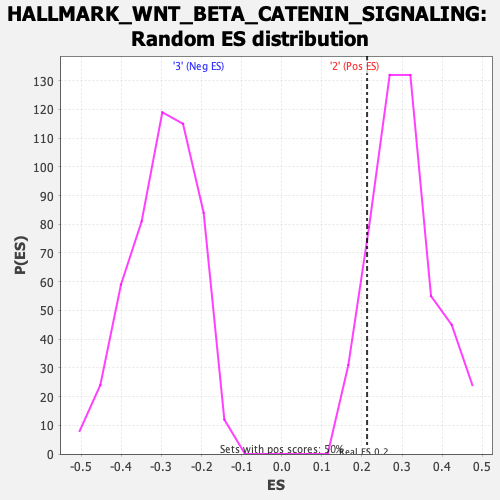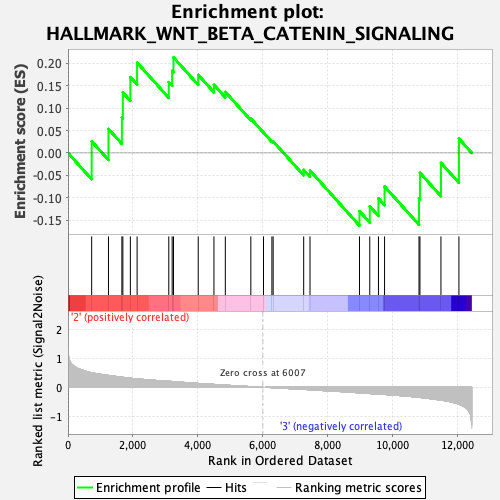
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.21287696 |
| Normalized Enrichment Score (NES) | 0.70085055 |
| Nominal p-value | 0.875502 |
| FDR q-value | 0.9886307 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ccnd2 | 729 | 0.500 | 0.0260 | Yes |
| 2 | Hdac11 | 1247 | 0.408 | 0.0535 | Yes |
| 3 | Notch1 | 1660 | 0.346 | 0.0788 | Yes |
| 4 | Cul1 | 1690 | 0.343 | 0.1346 | Yes |
| 5 | Dvl2 | 1921 | 0.312 | 0.1690 | Yes |
| 6 | Frat1 | 2129 | 0.289 | 0.2014 | Yes |
| 7 | Hdac2 | 3105 | 0.207 | 0.1579 | Yes |
| 8 | Hdac5 | 3209 | 0.198 | 0.1832 | Yes |
| 9 | Maml1 | 3250 | 0.194 | 0.2129 | Yes |
| 10 | Numb | 4012 | 0.133 | 0.1741 | No |
| 11 | Axin2 | 4496 | 0.098 | 0.1518 | No |
| 12 | Ncstn | 4847 | 0.073 | 0.1360 | No |
| 13 | Trp53 | 5633 | 0.023 | 0.0766 | No |
| 14 | Nkd1 | 6023 | 0.000 | 0.0453 | No |
| 15 | Ptch1 | 6282 | -0.011 | 0.0264 | No |
| 16 | Ncor2 | 6320 | -0.014 | 0.0258 | No |
| 17 | Psen2 | 7261 | -0.072 | -0.0377 | No |
| 18 | Ctnnb1 | 7456 | -0.083 | -0.0392 | No |
| 19 | Ppard | 8980 | -0.190 | -0.1297 | No |
| 20 | Adam17 | 9297 | -0.214 | -0.1188 | No |
| 21 | Rbpj | 9565 | -0.230 | -0.1013 | No |
| 22 | Axin1 | 9753 | -0.246 | -0.0748 | No |
| 23 | Csnk1e | 10814 | -0.347 | -0.1013 | No |
| 24 | Kat2a | 10843 | -0.350 | -0.0442 | No |
| 25 | Myc | 11490 | -0.438 | -0.0220 | No |
| 26 | Skp2 | 12044 | -0.582 | 0.0320 | No |