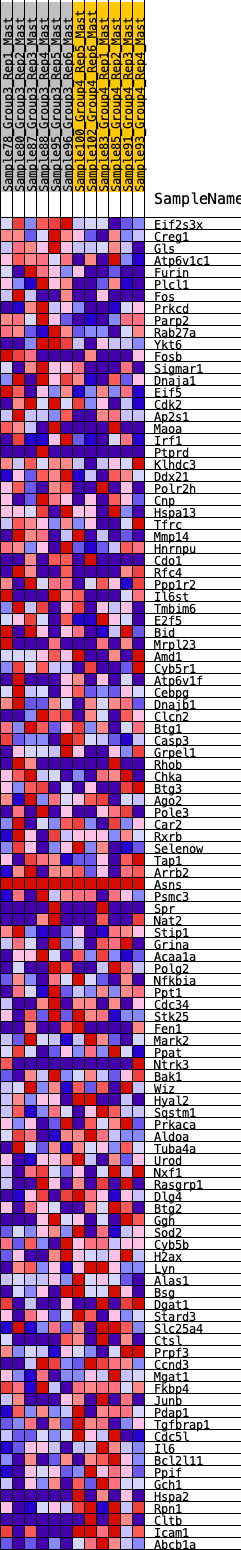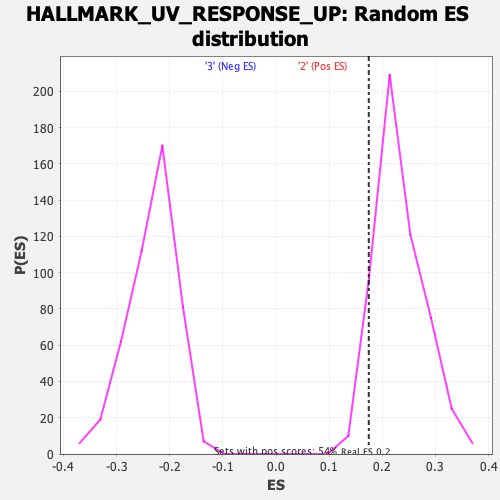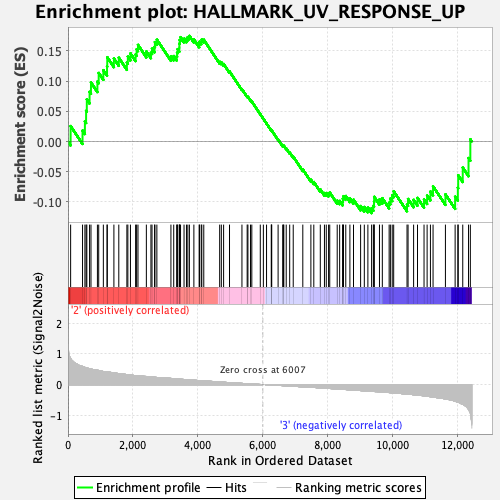
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.17465048 |
| Normalized Enrichment Score (NES) | 0.7500182 |
| Nominal p-value | 0.92449355 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Eif2s3x | 81 | 0.856 | 0.0256 | Yes |
| 2 | Creg1 | 448 | 0.591 | 0.0181 | Yes |
| 3 | Gls | 521 | 0.561 | 0.0333 | Yes |
| 4 | Atp6v1c1 | 558 | 0.547 | 0.0509 | Yes |
| 5 | Furin | 580 | 0.544 | 0.0696 | Yes |
| 6 | Plcl1 | 665 | 0.515 | 0.0821 | Yes |
| 7 | Fos | 708 | 0.504 | 0.0977 | Yes |
| 8 | Prkcd | 906 | 0.467 | 0.0992 | Yes |
| 9 | Parp2 | 946 | 0.462 | 0.1134 | Yes |
| 10 | Rab27a | 1089 | 0.428 | 0.1179 | Yes |
| 11 | Ykt6 | 1203 | 0.414 | 0.1243 | Yes |
| 12 | Fosb | 1210 | 0.412 | 0.1393 | Yes |
| 13 | Sigmar1 | 1414 | 0.385 | 0.1373 | Yes |
| 14 | Dnaja1 | 1565 | 0.358 | 0.1386 | Yes |
| 15 | Eif5 | 1814 | 0.325 | 0.1307 | Yes |
| 16 | Cdk2 | 1844 | 0.321 | 0.1404 | Yes |
| 17 | Ap2s1 | 1924 | 0.312 | 0.1457 | Yes |
| 18 | Maoa | 2083 | 0.294 | 0.1439 | Yes |
| 19 | Irf1 | 2114 | 0.291 | 0.1524 | Yes |
| 20 | Ptprd | 2156 | 0.285 | 0.1598 | Yes |
| 21 | Klhdc3 | 2414 | 0.265 | 0.1489 | Yes |
| 22 | Ddx21 | 2555 | 0.251 | 0.1470 | Yes |
| 23 | Polr2h | 2588 | 0.247 | 0.1537 | Yes |
| 24 | Cnp | 2667 | 0.239 | 0.1563 | Yes |
| 25 | Hspa13 | 2678 | 0.239 | 0.1645 | Yes |
| 26 | Tfrc | 2738 | 0.233 | 0.1684 | Yes |
| 27 | Mmp14 | 3171 | 0.202 | 0.1410 | Yes |
| 28 | Hnrnpu | 3257 | 0.194 | 0.1414 | Yes |
| 29 | Cdo1 | 3350 | 0.186 | 0.1409 | Yes |
| 30 | Rfc4 | 3361 | 0.185 | 0.1470 | Yes |
| 31 | Ppp1r2 | 3378 | 0.185 | 0.1527 | Yes |
| 32 | Il6st | 3431 | 0.180 | 0.1552 | Yes |
| 33 | Tmbim6 | 3432 | 0.179 | 0.1619 | Yes |
| 34 | E2f5 | 3442 | 0.178 | 0.1679 | Yes |
| 35 | Bid | 3467 | 0.175 | 0.1725 | Yes |
| 36 | Mrpl23 | 3575 | 0.167 | 0.1701 | Yes |
| 37 | Amd1 | 3650 | 0.161 | 0.1701 | Yes |
| 38 | Cyb5r1 | 3692 | 0.158 | 0.1727 | Yes |
| 39 | Atp6v1f | 3740 | 0.154 | 0.1747 | Yes |
| 40 | Cebpg | 3878 | 0.144 | 0.1689 | No |
| 41 | Dnajb1 | 4042 | 0.130 | 0.1606 | No |
| 42 | Clcn2 | 4049 | 0.130 | 0.1650 | No |
| 43 | Btg1 | 4104 | 0.126 | 0.1654 | No |
| 44 | Casp3 | 4122 | 0.125 | 0.1687 | No |
| 45 | Grpel1 | 4183 | 0.121 | 0.1683 | No |
| 46 | Rhob | 4673 | 0.085 | 0.1319 | No |
| 47 | Chka | 4724 | 0.082 | 0.1309 | No |
| 48 | Btg3 | 4794 | 0.078 | 0.1282 | No |
| 49 | Ago2 | 4976 | 0.065 | 0.1160 | No |
| 50 | Pole3 | 5359 | 0.040 | 0.0865 | No |
| 51 | Car2 | 5520 | 0.030 | 0.0746 | No |
| 52 | Rxrb | 5543 | 0.028 | 0.0739 | No |
| 53 | Selenow | 5623 | 0.023 | 0.0684 | No |
| 54 | Tap1 | 5663 | 0.021 | 0.0660 | No |
| 55 | Arrb2 | 5924 | 0.005 | 0.0451 | No |
| 56 | Asns | 6019 | 0.000 | 0.0375 | No |
| 57 | Psmc3 | 6117 | -0.002 | 0.0297 | No |
| 58 | Spr | 6262 | -0.010 | 0.0184 | No |
| 59 | Nat2 | 6273 | -0.011 | 0.0180 | No |
| 60 | Stip1 | 6274 | -0.011 | 0.0184 | No |
| 61 | Grina | 6473 | -0.025 | 0.0032 | No |
| 62 | Acaa1a | 6615 | -0.032 | -0.0070 | No |
| 63 | Polg2 | 6637 | -0.034 | -0.0074 | No |
| 64 | Nfkbia | 6649 | -0.035 | -0.0070 | No |
| 65 | Ppt1 | 6723 | -0.040 | -0.0114 | No |
| 66 | Cdc34 | 6830 | -0.046 | -0.0183 | No |
| 67 | Stk25 | 6939 | -0.052 | -0.0251 | No |
| 68 | Fen1 | 7234 | -0.070 | -0.0463 | No |
| 69 | Mark2 | 7483 | -0.085 | -0.0632 | No |
| 70 | Ppat | 7573 | -0.091 | -0.0670 | No |
| 71 | Ntrk3 | 7773 | -0.105 | -0.0792 | No |
| 72 | Bak1 | 7901 | -0.115 | -0.0852 | No |
| 73 | Wiz | 7957 | -0.118 | -0.0852 | No |
| 74 | Hyal2 | 8028 | -0.123 | -0.0863 | No |
| 75 | Sqstm1 | 8064 | -0.126 | -0.0844 | No |
| 76 | Prkaca | 8294 | -0.140 | -0.0977 | No |
| 77 | Aldoa | 8369 | -0.146 | -0.0982 | No |
| 78 | Tuba4a | 8462 | -0.152 | -0.1000 | No |
| 79 | Urod | 8468 | -0.152 | -0.0947 | No |
| 80 | Nxf1 | 8488 | -0.153 | -0.0905 | No |
| 81 | Rasgrp1 | 8558 | -0.159 | -0.0901 | No |
| 82 | Dlg4 | 8685 | -0.170 | -0.0939 | No |
| 83 | Btg2 | 8796 | -0.177 | -0.0962 | No |
| 84 | Ggh | 9015 | -0.193 | -0.1067 | No |
| 85 | Sod2 | 9129 | -0.201 | -0.1083 | No |
| 86 | Cyb5b | 9239 | -0.210 | -0.1092 | No |
| 87 | H2ax | 9354 | -0.218 | -0.1103 | No |
| 88 | Lyn | 9408 | -0.222 | -0.1063 | No |
| 89 | Alas1 | 9429 | -0.223 | -0.0995 | No |
| 90 | Bsg | 9434 | -0.224 | -0.0914 | No |
| 91 | Dgat1 | 9596 | -0.233 | -0.0957 | No |
| 92 | Stard3 | 9684 | -0.240 | -0.0938 | No |
| 93 | Slc25a4 | 9894 | -0.258 | -0.1010 | No |
| 94 | Ctsl | 9932 | -0.261 | -0.0942 | No |
| 95 | Prpf3 | 9989 | -0.266 | -0.0888 | No |
| 96 | Ccnd3 | 10034 | -0.270 | -0.0822 | No |
| 97 | Mgat1 | 10445 | -0.304 | -0.1041 | No |
| 98 | Fkbp4 | 10477 | -0.308 | -0.0950 | No |
| 99 | Junb | 10648 | -0.329 | -0.0965 | No |
| 100 | Pdap1 | 10768 | -0.342 | -0.0933 | No |
| 101 | Tgfbrap1 | 10969 | -0.364 | -0.0958 | No |
| 102 | Cdc5l | 11064 | -0.379 | -0.0892 | No |
| 103 | Il6 | 11166 | -0.392 | -0.0827 | No |
| 104 | Bcl2l11 | 11247 | -0.403 | -0.0741 | No |
| 105 | Ppif | 11628 | -0.467 | -0.0873 | No |
| 106 | Gch1 | 11927 | -0.543 | -0.0911 | No |
| 107 | Hspa2 | 12011 | -0.569 | -0.0765 | No |
| 108 | Rpn1 | 12022 | -0.572 | -0.0558 | No |
| 109 | Cltb | 12162 | -0.638 | -0.0431 | No |
| 110 | Icam1 | 12340 | -0.803 | -0.0273 | No |
| 111 | Abcb1a | 12396 | -0.944 | 0.0036 | No |