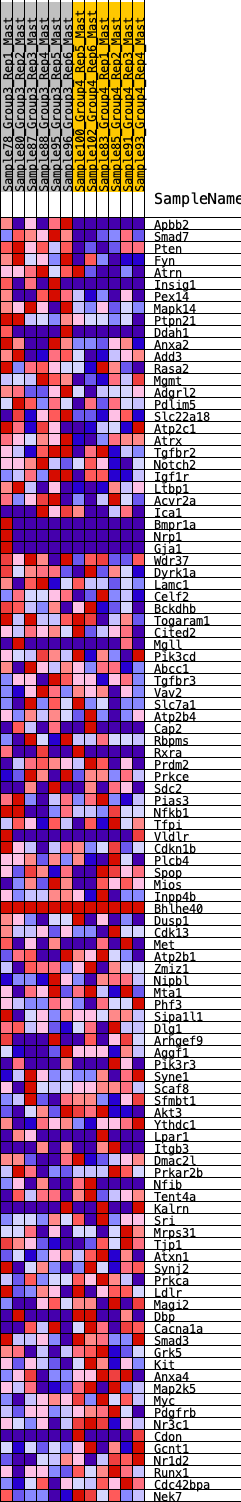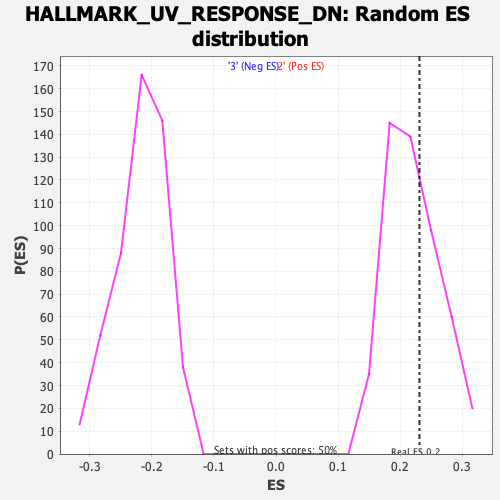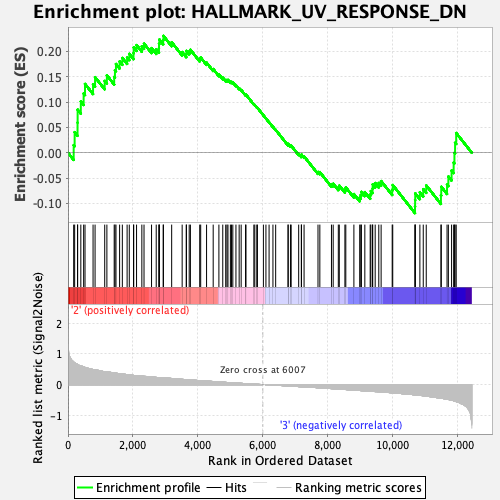
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.23082669 |
| Normalized Enrichment Score (NES) | 1.0455182 |
| Nominal p-value | 0.38229376 |
| FDR q-value | 0.70655024 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Apbb2 | 175 | 0.736 | 0.0149 | Yes |
| 2 | Smad7 | 205 | 0.710 | 0.0406 | Yes |
| 3 | Pten | 294 | 0.656 | 0.0594 | Yes |
| 4 | Fyn | 297 | 0.653 | 0.0850 | Yes |
| 5 | Atrn | 395 | 0.609 | 0.1012 | Yes |
| 6 | Insig1 | 482 | 0.577 | 0.1170 | Yes |
| 7 | Pex14 | 524 | 0.560 | 0.1359 | Yes |
| 8 | Mapk14 | 772 | 0.491 | 0.1353 | Yes |
| 9 | Ptpn21 | 834 | 0.478 | 0.1492 | Yes |
| 10 | Ddah1 | 1131 | 0.421 | 0.1418 | Yes |
| 11 | Anxa2 | 1198 | 0.415 | 0.1529 | Yes |
| 12 | Add3 | 1423 | 0.383 | 0.1499 | Yes |
| 13 | Rasa2 | 1447 | 0.379 | 0.1630 | Yes |
| 14 | Mgmt | 1478 | 0.373 | 0.1753 | Yes |
| 15 | Adgrl2 | 1590 | 0.354 | 0.1803 | Yes |
| 16 | Pdlim5 | 1674 | 0.344 | 0.1872 | Yes |
| 17 | Slc22a18 | 1821 | 0.324 | 0.1882 | Yes |
| 18 | Atp2c1 | 1888 | 0.317 | 0.1953 | Yes |
| 19 | Atrx | 2018 | 0.300 | 0.1967 | Yes |
| 20 | Tgfbr2 | 2025 | 0.299 | 0.2080 | Yes |
| 21 | Notch2 | 2112 | 0.292 | 0.2126 | Yes |
| 22 | Igf1r | 2275 | 0.274 | 0.2103 | Yes |
| 23 | Ltbp1 | 2342 | 0.268 | 0.2155 | Yes |
| 24 | Acvr2a | 2574 | 0.249 | 0.2066 | Yes |
| 25 | Ica1 | 2718 | 0.234 | 0.2043 | Yes |
| 26 | Bmpr1a | 2803 | 0.227 | 0.2065 | Yes |
| 27 | Nrp1 | 2805 | 0.227 | 0.2154 | Yes |
| 28 | Gja1 | 2815 | 0.227 | 0.2237 | Yes |
| 29 | Wdr37 | 2933 | 0.217 | 0.2228 | Yes |
| 30 | Dyrk1a | 2940 | 0.217 | 0.2308 | Yes |
| 31 | Lamc1 | 3194 | 0.200 | 0.2182 | No |
| 32 | Celf2 | 3517 | 0.171 | 0.1989 | No |
| 33 | Bckdhb | 3644 | 0.161 | 0.1951 | No |
| 34 | Togaram1 | 3648 | 0.161 | 0.2012 | No |
| 35 | Cited2 | 3734 | 0.154 | 0.2004 | No |
| 36 | Mgll | 3771 | 0.151 | 0.2034 | No |
| 37 | Pik3cd | 4058 | 0.129 | 0.1853 | No |
| 38 | Abcc1 | 4088 | 0.128 | 0.1880 | No |
| 39 | Tgfbr3 | 4269 | 0.113 | 0.1779 | No |
| 40 | Vav2 | 4473 | 0.100 | 0.1654 | No |
| 41 | Slc7a1 | 4649 | 0.088 | 0.1547 | No |
| 42 | Atp2b4 | 4768 | 0.080 | 0.1483 | No |
| 43 | Cap2 | 4859 | 0.073 | 0.1438 | No |
| 44 | Rbpms | 4897 | 0.070 | 0.1436 | No |
| 45 | Rxra | 4928 | 0.068 | 0.1438 | No |
| 46 | Prdm2 | 5003 | 0.063 | 0.1403 | No |
| 47 | Prkce | 5038 | 0.061 | 0.1400 | No |
| 48 | Sdc2 | 5078 | 0.058 | 0.1391 | No |
| 49 | Pias3 | 5174 | 0.051 | 0.1334 | No |
| 50 | Nfkb1 | 5273 | 0.046 | 0.1273 | No |
| 51 | Tfpi | 5335 | 0.041 | 0.1240 | No |
| 52 | Vldlr | 5470 | 0.033 | 0.1145 | No |
| 53 | Cdkn1b | 5483 | 0.033 | 0.1148 | No |
| 54 | Plcb4 | 5726 | 0.018 | 0.0959 | No |
| 55 | Spop | 5755 | 0.016 | 0.0942 | No |
| 56 | Mios | 5817 | 0.012 | 0.0898 | No |
| 57 | Inpp4b | 5836 | 0.011 | 0.0887 | No |
| 58 | Bhlhe40 | 6020 | 0.000 | 0.0739 | No |
| 59 | Dusp1 | 6098 | -0.001 | 0.0677 | No |
| 60 | Cdk13 | 6195 | -0.006 | 0.0602 | No |
| 61 | Met | 6314 | -0.014 | 0.0512 | No |
| 62 | Atp2b1 | 6398 | -0.019 | 0.0452 | No |
| 63 | Zmiz1 | 6780 | -0.043 | 0.0160 | No |
| 64 | Nipbl | 6783 | -0.043 | 0.0175 | No |
| 65 | Mta1 | 6854 | -0.048 | 0.0137 | No |
| 66 | Phf3 | 6875 | -0.049 | 0.0140 | No |
| 67 | Sipa1l1 | 7107 | -0.063 | -0.0022 | No |
| 68 | Dlg1 | 7188 | -0.067 | -0.0060 | No |
| 69 | Arhgef9 | 7194 | -0.068 | -0.0037 | No |
| 70 | Aggf1 | 7273 | -0.073 | -0.0072 | No |
| 71 | Pik3r3 | 7702 | -0.100 | -0.0379 | No |
| 72 | Syne1 | 7759 | -0.105 | -0.0383 | No |
| 73 | Scaf8 | 8117 | -0.129 | -0.0622 | No |
| 74 | Sfmbt1 | 8170 | -0.132 | -0.0612 | No |
| 75 | Akt3 | 8328 | -0.143 | -0.0682 | No |
| 76 | Ythdc1 | 8358 | -0.145 | -0.0649 | No |
| 77 | Lpar1 | 8531 | -0.156 | -0.0726 | No |
| 78 | Itgb3 | 8559 | -0.160 | -0.0685 | No |
| 79 | Dmac2l | 8808 | -0.177 | -0.0816 | No |
| 80 | Prkar2b | 8987 | -0.191 | -0.0885 | No |
| 81 | Nfib | 9020 | -0.193 | -0.0835 | No |
| 82 | Tent4a | 9040 | -0.195 | -0.0773 | No |
| 83 | Kalrn | 9145 | -0.202 | -0.0777 | No |
| 84 | Sri | 9310 | -0.215 | -0.0825 | No |
| 85 | Mrps31 | 9331 | -0.217 | -0.0756 | No |
| 86 | Tjp1 | 9383 | -0.220 | -0.0710 | No |
| 87 | Atxn1 | 9387 | -0.220 | -0.0626 | No |
| 88 | Synj2 | 9464 | -0.226 | -0.0598 | No |
| 89 | Prkca | 9575 | -0.231 | -0.0596 | No |
| 90 | Ldlr | 9647 | -0.237 | -0.0560 | No |
| 91 | Magi2 | 9988 | -0.266 | -0.0730 | No |
| 92 | Dbp | 10005 | -0.267 | -0.0638 | No |
| 93 | Cacna1a | 10691 | -0.332 | -0.1062 | No |
| 94 | Smad3 | 10695 | -0.333 | -0.0933 | No |
| 95 | Grk5 | 10699 | -0.334 | -0.0803 | No |
| 96 | Kit | 10841 | -0.350 | -0.0779 | No |
| 97 | Anxa4 | 10945 | -0.361 | -0.0720 | No |
| 98 | Map2k5 | 11037 | -0.374 | -0.0646 | No |
| 99 | Myc | 11490 | -0.438 | -0.0839 | No |
| 100 | Pdgfrb | 11500 | -0.441 | -0.0672 | No |
| 101 | Nr3c1 | 11678 | -0.476 | -0.0628 | No |
| 102 | Cdon | 11719 | -0.483 | -0.0469 | No |
| 103 | Gcnt1 | 11818 | -0.500 | -0.0351 | No |
| 104 | Nr1d2 | 11882 | -0.527 | -0.0194 | No |
| 105 | Runx1 | 11906 | -0.537 | -0.0001 | No |
| 106 | Cdc42bpa | 11926 | -0.543 | 0.0198 | No |
| 107 | Nek7 | 11960 | -0.552 | 0.0390 | No |