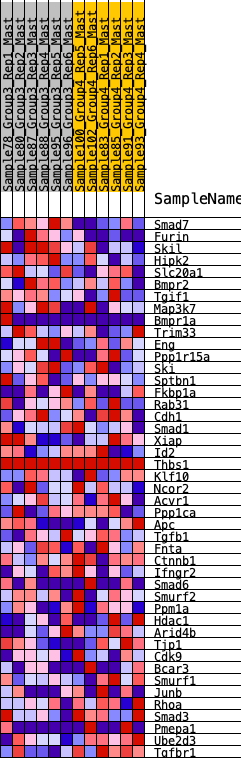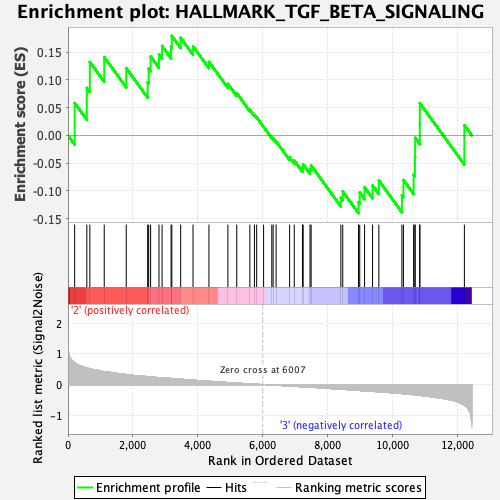
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.17926991 |
| Normalized Enrichment Score (NES) | 0.7297783 |
| Nominal p-value | 0.904943 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smad7 | 205 | 0.710 | 0.0584 | Yes |
| 2 | Furin | 580 | 0.544 | 0.0856 | Yes |
| 3 | Skil | 673 | 0.513 | 0.1323 | Yes |
| 4 | Hipk2 | 1116 | 0.422 | 0.1412 | Yes |
| 5 | Slc20a1 | 1794 | 0.328 | 0.1212 | Yes |
| 6 | Bmpr2 | 2454 | 0.261 | 0.0955 | Yes |
| 7 | Tgif1 | 2485 | 0.258 | 0.1204 | Yes |
| 8 | Map3k7 | 2545 | 0.252 | 0.1422 | Yes |
| 9 | Bmpr1a | 2803 | 0.227 | 0.1455 | Yes |
| 10 | Trim33 | 2898 | 0.220 | 0.1611 | Yes |
| 11 | Eng | 3176 | 0.201 | 0.1600 | Yes |
| 12 | Ppp1r15a | 3199 | 0.199 | 0.1793 | Yes |
| 13 | Ski | 3471 | 0.175 | 0.1759 | No |
| 14 | Sptbn1 | 3852 | 0.146 | 0.1606 | No |
| 15 | Fkbp1a | 4343 | 0.109 | 0.1326 | No |
| 16 | Rab31 | 4926 | 0.068 | 0.0928 | No |
| 17 | Cdh1 | 5199 | 0.049 | 0.0761 | No |
| 18 | Smad1 | 5603 | 0.024 | 0.0461 | No |
| 19 | Xiap | 5744 | 0.016 | 0.0366 | No |
| 20 | Id2 | 5813 | 0.012 | 0.0324 | No |
| 21 | Thbs1 | 6026 | 0.000 | 0.0153 | No |
| 22 | Klf10 | 6279 | -0.011 | -0.0039 | No |
| 23 | Ncor2 | 6320 | -0.014 | -0.0056 | No |
| 24 | Acvr1 | 6411 | -0.020 | -0.0108 | No |
| 25 | Ppp1ca | 6828 | -0.046 | -0.0395 | No |
| 26 | Apc | 6971 | -0.054 | -0.0452 | No |
| 27 | Tgfb1 | 7224 | -0.069 | -0.0582 | No |
| 28 | Fnta | 7245 | -0.071 | -0.0523 | No |
| 29 | Ctnnb1 | 7456 | -0.083 | -0.0605 | No |
| 30 | Ifngr2 | 7491 | -0.086 | -0.0542 | No |
| 31 | Smad6 | 8407 | -0.148 | -0.1124 | No |
| 32 | Smurf2 | 8465 | -0.152 | -0.1010 | No |
| 33 | Ppm1a | 8950 | -0.188 | -0.1202 | No |
| 34 | Hdac1 | 8988 | -0.191 | -0.1030 | No |
| 35 | Arid4b | 9137 | -0.201 | -0.0937 | No |
| 36 | Tjp1 | 9383 | -0.220 | -0.0902 | No |
| 37 | Cdk9 | 9577 | -0.232 | -0.0813 | No |
| 38 | Bcar3 | 10287 | -0.292 | -0.1077 | No |
| 39 | Smurf1 | 10334 | -0.296 | -0.0802 | No |
| 40 | Junb | 10648 | -0.329 | -0.0708 | No |
| 41 | Rhoa | 10688 | -0.332 | -0.0389 | No |
| 42 | Smad3 | 10695 | -0.333 | -0.0043 | No |
| 43 | Pmepa1 | 10838 | -0.350 | 0.0212 | No |
| 44 | Ube2d3 | 10840 | -0.350 | 0.0580 | No |
| 45 | Tgfbr1 | 12212 | -0.673 | 0.0185 | No |