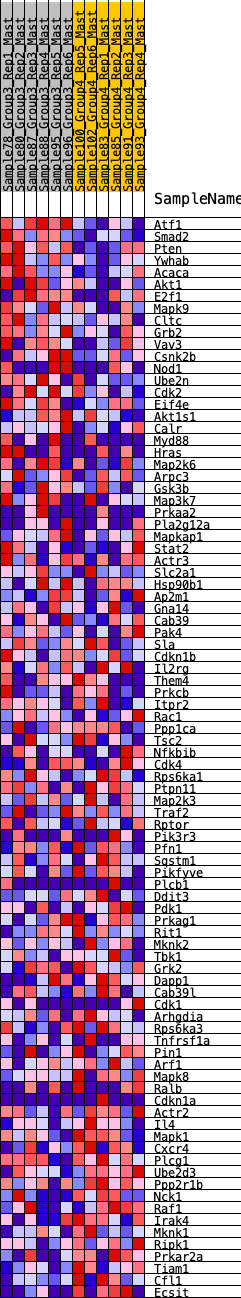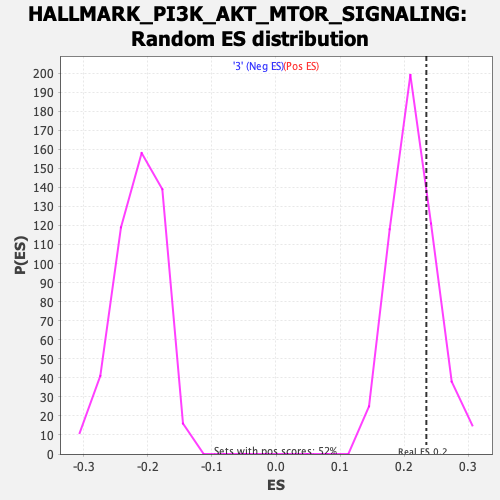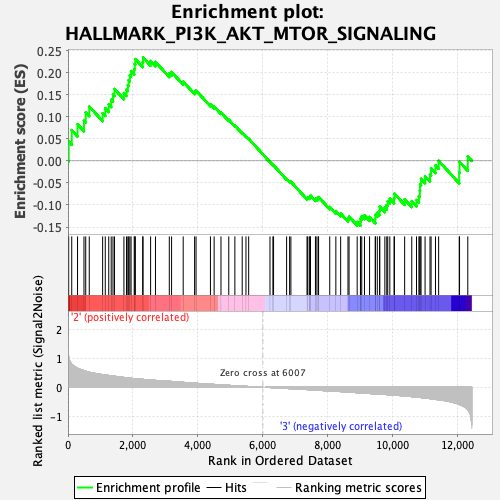
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.23440231 |
| Normalized Enrichment Score (NES) | 1.0971054 |
| Nominal p-value | 0.25387597 |
| FDR q-value | 0.6757773 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atf1 | 21 | 1.078 | 0.0438 | Yes |
| 2 | Smad2 | 115 | 0.792 | 0.0697 | Yes |
| 3 | Pten | 294 | 0.656 | 0.0829 | Yes |
| 4 | Ywhab | 492 | 0.572 | 0.0911 | Yes |
| 5 | Acaca | 546 | 0.550 | 0.1100 | Yes |
| 6 | Akt1 | 655 | 0.518 | 0.1231 | Yes |
| 7 | E2f1 | 1067 | 0.431 | 0.1080 | Yes |
| 8 | Mapk9 | 1148 | 0.419 | 0.1192 | Yes |
| 9 | Cltc | 1255 | 0.407 | 0.1278 | Yes |
| 10 | Grb2 | 1331 | 0.395 | 0.1384 | Yes |
| 11 | Vav3 | 1386 | 0.387 | 0.1504 | Yes |
| 12 | Csnk2b | 1432 | 0.381 | 0.1629 | Yes |
| 13 | Nod1 | 1723 | 0.339 | 0.1537 | Yes |
| 14 | Ube2n | 1803 | 0.327 | 0.1611 | Yes |
| 15 | Cdk2 | 1844 | 0.321 | 0.1714 | Yes |
| 16 | Eif4e | 1869 | 0.319 | 0.1829 | Yes |
| 17 | Akt1s1 | 1906 | 0.314 | 0.1932 | Yes |
| 18 | Calr | 1943 | 0.311 | 0.2034 | Yes |
| 19 | Myd88 | 2040 | 0.298 | 0.2082 | Yes |
| 20 | Hras | 2056 | 0.297 | 0.2195 | Yes |
| 21 | Map2k6 | 2075 | 0.295 | 0.2305 | Yes |
| 22 | Arpc3 | 2301 | 0.272 | 0.2237 | Yes |
| 23 | Gsk3b | 2312 | 0.272 | 0.2344 | Yes |
| 24 | Map3k7 | 2545 | 0.252 | 0.2263 | No |
| 25 | Prkaa2 | 2695 | 0.236 | 0.2242 | No |
| 26 | Pla2g12a | 3121 | 0.206 | 0.1985 | No |
| 27 | Mapkap1 | 3191 | 0.200 | 0.2013 | No |
| 28 | Stat2 | 3549 | 0.169 | 0.1795 | No |
| 29 | Actr3 | 3900 | 0.142 | 0.1572 | No |
| 30 | Slc2a1 | 3945 | 0.138 | 0.1595 | No |
| 31 | Hsp90b1 | 4388 | 0.107 | 0.1282 | No |
| 32 | Ap2m1 | 4502 | 0.098 | 0.1232 | No |
| 33 | Gna14 | 4713 | 0.082 | 0.1096 | No |
| 34 | Cab39 | 4953 | 0.067 | 0.0931 | No |
| 35 | Pak4 | 5141 | 0.053 | 0.0802 | No |
| 36 | Sla | 5367 | 0.039 | 0.0636 | No |
| 37 | Cdkn1b | 5483 | 0.033 | 0.0557 | No |
| 38 | Il2rg | 5566 | 0.027 | 0.0502 | No |
| 39 | Them4 | 6223 | -0.008 | -0.0026 | No |
| 40 | Prkcb | 6313 | -0.013 | -0.0092 | No |
| 41 | Itpr2 | 6333 | -0.015 | -0.0101 | No |
| 42 | Rac1 | 6735 | -0.040 | -0.0409 | No |
| 43 | Ppp1ca | 6828 | -0.046 | -0.0464 | No |
| 44 | Tsc2 | 6867 | -0.048 | -0.0474 | No |
| 45 | Nfkbib | 7364 | -0.078 | -0.0843 | No |
| 46 | Cdk4 | 7384 | -0.079 | -0.0825 | No |
| 47 | Rps6ka1 | 7434 | -0.083 | -0.0830 | No |
| 48 | Ptpn11 | 7455 | -0.083 | -0.0811 | No |
| 49 | Map2k3 | 7471 | -0.084 | -0.0787 | No |
| 50 | Traf2 | 7623 | -0.094 | -0.0870 | No |
| 51 | Rptor | 7644 | -0.096 | -0.0846 | No |
| 52 | Pik3r3 | 7702 | -0.100 | -0.0849 | No |
| 53 | Pfn1 | 7720 | -0.102 | -0.0820 | No |
| 54 | Sqstm1 | 8064 | -0.126 | -0.1045 | No |
| 55 | Pikfyve | 8252 | -0.137 | -0.1138 | No |
| 56 | Plcb1 | 8400 | -0.147 | -0.1195 | No |
| 57 | Ddit3 | 8626 | -0.165 | -0.1308 | No |
| 58 | Pdk1 | 8659 | -0.168 | -0.1263 | No |
| 59 | Prkag1 | 8909 | -0.185 | -0.1387 | No |
| 60 | Rit1 | 9010 | -0.193 | -0.1386 | No |
| 61 | Mknk2 | 9013 | -0.193 | -0.1307 | No |
| 62 | Tbk1 | 9050 | -0.195 | -0.1253 | No |
| 63 | Grk2 | 9134 | -0.201 | -0.1235 | No |
| 64 | Dapp1 | 9291 | -0.214 | -0.1272 | No |
| 65 | Cab39l | 9468 | -0.226 | -0.1319 | No |
| 66 | Cdk1 | 9469 | -0.226 | -0.1223 | No |
| 67 | Arhgdia | 9529 | -0.227 | -0.1175 | No |
| 68 | Rps6ka3 | 9599 | -0.234 | -0.1132 | No |
| 69 | Tnfrsf1a | 9605 | -0.234 | -0.1038 | No |
| 70 | Pin1 | 9762 | -0.246 | -0.1060 | No |
| 71 | Arf1 | 9821 | -0.251 | -0.1001 | No |
| 72 | Mapk8 | 9851 | -0.255 | -0.0917 | No |
| 73 | Ralb | 9915 | -0.260 | -0.0858 | No |
| 74 | Cdkn1a | 10046 | -0.270 | -0.0849 | No |
| 75 | Actr2 | 10057 | -0.271 | -0.0743 | No |
| 76 | Il4 | 10372 | -0.298 | -0.0872 | No |
| 77 | Mapk1 | 10591 | -0.321 | -0.0913 | No |
| 78 | Cxcr4 | 10737 | -0.338 | -0.0888 | No |
| 79 | Plcg1 | 10811 | -0.347 | -0.0801 | No |
| 80 | Ube2d3 | 10840 | -0.350 | -0.0676 | No |
| 81 | Ppp2r1b | 10847 | -0.350 | -0.0533 | No |
| 82 | Nck1 | 10877 | -0.354 | -0.0407 | No |
| 83 | Raf1 | 11001 | -0.369 | -0.0351 | No |
| 84 | Irak4 | 11153 | -0.390 | -0.0309 | No |
| 85 | Mknk1 | 11186 | -0.395 | -0.0168 | No |
| 86 | Ripk1 | 11322 | -0.416 | -0.0102 | No |
| 87 | Prkar2a | 11419 | -0.430 | 0.0001 | No |
| 88 | Tiam1 | 12053 | -0.585 | -0.0264 | No |
| 89 | Cfl1 | 12063 | -0.592 | -0.0022 | No |
| 90 | Ecsit | 12317 | -0.776 | 0.0100 | No |