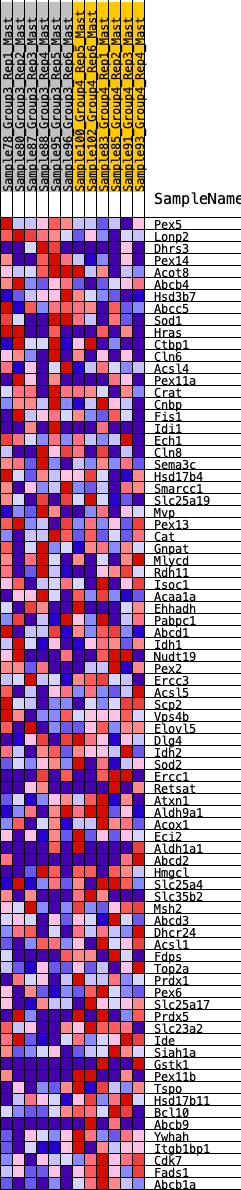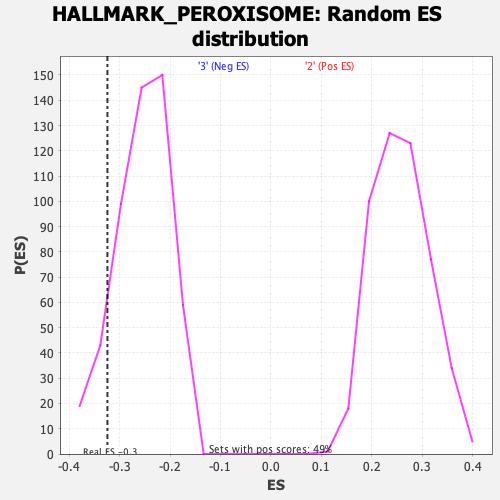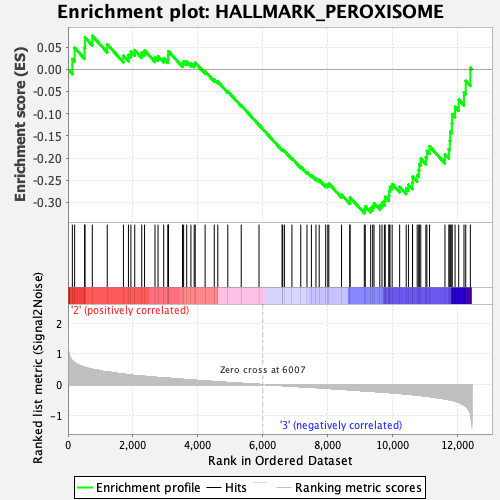
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.32410643 |
| Normalized Enrichment Score (NES) | -1.2769907 |
| Nominal p-value | 0.10485437 |
| FDR q-value | 0.95599526 |
| FWER p-Value | 0.843 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 134 | 0.777 | 0.0230 | No |
| 2 | Lonp2 | 204 | 0.712 | 0.0485 | No |
| 3 | Dhrs3 | 511 | 0.564 | 0.0483 | No |
| 4 | Pex14 | 524 | 0.560 | 0.0718 | No |
| 5 | Acot8 | 749 | 0.496 | 0.0753 | No |
| 6 | Abcb4 | 1209 | 0.413 | 0.0561 | No |
| 7 | Hsd3b7 | 1708 | 0.340 | 0.0307 | No |
| 8 | Abcc5 | 1862 | 0.320 | 0.0322 | No |
| 9 | Sod1 | 1938 | 0.311 | 0.0397 | No |
| 10 | Hras | 2056 | 0.297 | 0.0432 | No |
| 11 | Ctbp1 | 2272 | 0.274 | 0.0377 | No |
| 12 | Cln6 | 2358 | 0.268 | 0.0425 | No |
| 13 | Acsl4 | 2680 | 0.238 | 0.0269 | No |
| 14 | Pex11a | 2774 | 0.230 | 0.0295 | No |
| 15 | Crat | 2949 | 0.216 | 0.0248 | No |
| 16 | Cnbp | 3078 | 0.210 | 0.0236 | No |
| 17 | Fis1 | 3089 | 0.208 | 0.0318 | No |
| 18 | Idi1 | 3094 | 0.208 | 0.0406 | No |
| 19 | Ech1 | 3529 | 0.170 | 0.0129 | No |
| 20 | Cln8 | 3562 | 0.168 | 0.0176 | No |
| 21 | Sema3c | 3657 | 0.160 | 0.0170 | No |
| 22 | Hsd17b4 | 3787 | 0.150 | 0.0131 | No |
| 23 | Smarcc1 | 3888 | 0.143 | 0.0112 | No |
| 24 | Slc25a19 | 3919 | 0.140 | 0.0149 | No |
| 25 | Mvp | 4224 | 0.117 | -0.0046 | No |
| 26 | Pex13 | 4506 | 0.098 | -0.0231 | No |
| 27 | Cat | 4615 | 0.090 | -0.0279 | No |
| 28 | Gnpat | 4923 | 0.068 | -0.0497 | No |
| 29 | Mlycd | 5337 | 0.041 | -0.0814 | No |
| 30 | Rdh11 | 5885 | 0.007 | -0.1253 | No |
| 31 | Isoc1 | 6595 | -0.031 | -0.1813 | No |
| 32 | Acaa1a | 6615 | -0.032 | -0.1815 | No |
| 33 | Ehhadh | 6667 | -0.036 | -0.1840 | No |
| 34 | Pabpc1 | 6899 | -0.050 | -0.2005 | No |
| 35 | Abcd1 | 7171 | -0.067 | -0.2195 | No |
| 36 | Idh1 | 7362 | -0.078 | -0.2315 | No |
| 37 | Nudt19 | 7499 | -0.086 | -0.2387 | No |
| 38 | Pex2 | 7639 | -0.096 | -0.2458 | No |
| 39 | Ercc3 | 7739 | -0.103 | -0.2493 | No |
| 40 | Acsl5 | 7940 | -0.117 | -0.2604 | No |
| 41 | Scp2 | 8004 | -0.121 | -0.2602 | No |
| 42 | Vps4b | 8036 | -0.124 | -0.2573 | No |
| 43 | Elovl5 | 8427 | -0.149 | -0.2824 | No |
| 44 | Dlg4 | 8685 | -0.170 | -0.2958 | No |
| 45 | Idh2 | 8688 | -0.170 | -0.2885 | No |
| 46 | Sod2 | 9129 | -0.201 | -0.3153 | Yes |
| 47 | Ercc1 | 9158 | -0.203 | -0.3088 | Yes |
| 48 | Retsat | 9325 | -0.216 | -0.3128 | Yes |
| 49 | Atxn1 | 9387 | -0.220 | -0.3081 | Yes |
| 50 | Aldh9a1 | 9428 | -0.223 | -0.3016 | Yes |
| 51 | Acox1 | 9607 | -0.234 | -0.3058 | Yes |
| 52 | Eci2 | 9674 | -0.240 | -0.3007 | Yes |
| 53 | Aldh1a1 | 9748 | -0.245 | -0.2959 | Yes |
| 54 | Abcd2 | 9773 | -0.247 | -0.2871 | Yes |
| 55 | Hmgcl | 9886 | -0.257 | -0.2849 | Yes |
| 56 | Slc25a4 | 9894 | -0.258 | -0.2742 | Yes |
| 57 | Slc35b2 | 9920 | -0.260 | -0.2649 | Yes |
| 58 | Msh2 | 9987 | -0.266 | -0.2587 | Yes |
| 59 | Abcd3 | 10218 | -0.285 | -0.2649 | Yes |
| 60 | Dhcr24 | 10419 | -0.302 | -0.2679 | Yes |
| 61 | Acsl1 | 10488 | -0.310 | -0.2599 | Yes |
| 62 | Fdps | 10613 | -0.324 | -0.2558 | Yes |
| 63 | Top2a | 10616 | -0.324 | -0.2418 | Yes |
| 64 | Prdx1 | 10758 | -0.341 | -0.2384 | Yes |
| 65 | Pex6 | 10807 | -0.346 | -0.2272 | Yes |
| 66 | Slc25a17 | 10824 | -0.348 | -0.2133 | Yes |
| 67 | Prdx5 | 10869 | -0.353 | -0.2015 | Yes |
| 68 | Slc23a2 | 11026 | -0.372 | -0.1979 | Yes |
| 69 | Ide | 11057 | -0.376 | -0.1839 | Yes |
| 70 | Siah1a | 11138 | -0.389 | -0.1734 | Yes |
| 71 | Gstk1 | 11614 | -0.465 | -0.1916 | Yes |
| 72 | Pex11b | 11733 | -0.487 | -0.1799 | Yes |
| 73 | Tspo | 11763 | -0.493 | -0.1608 | Yes |
| 74 | Hsd17b11 | 11777 | -0.495 | -0.1402 | Yes |
| 75 | Bcl10 | 11826 | -0.503 | -0.1222 | Yes |
| 76 | Abcb9 | 11835 | -0.506 | -0.1008 | Yes |
| 77 | Ywhah | 11925 | -0.543 | -0.0843 | Yes |
| 78 | Itgb1bp1 | 12037 | -0.578 | -0.0681 | Yes |
| 79 | Cdk7 | 12200 | -0.663 | -0.0523 | Yes |
| 80 | Fads1 | 12254 | -0.700 | -0.0261 | Yes |
| 81 | Abcb1a | 12396 | -0.944 | 0.0036 | Yes |