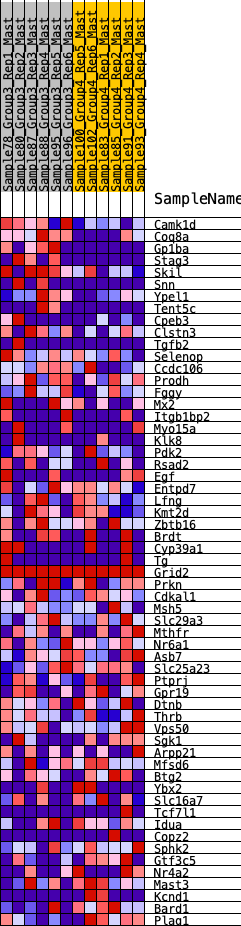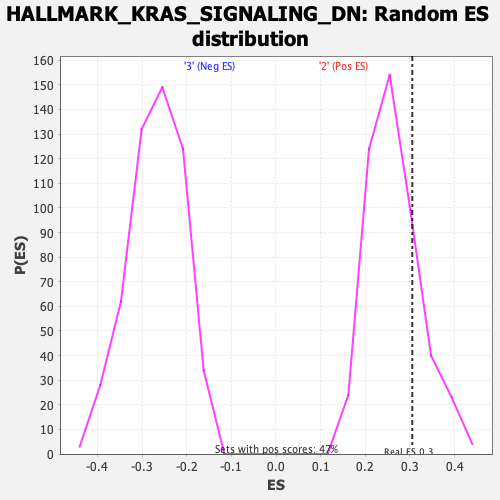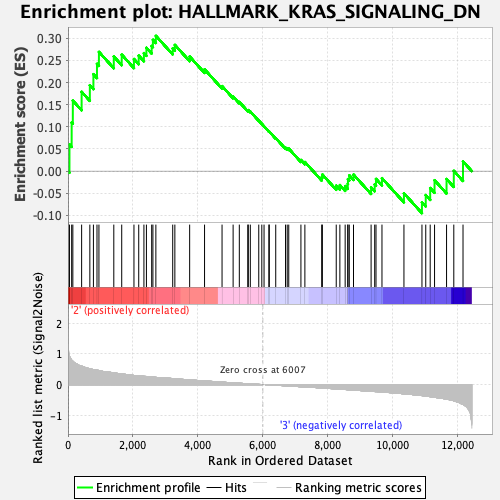
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.30517334 |
| Normalized Enrichment Score (NES) | 1.1548306 |
| Nominal p-value | 0.1965812 |
| FDR q-value | 0.5628335 |
| FWER p-Value | 0.971 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Camk1d | 45 | 0.926 | 0.0601 | Yes |
| 2 | Coq8a | 113 | 0.798 | 0.1096 | Yes |
| 3 | Gp1ba | 149 | 0.759 | 0.1590 | Yes |
| 4 | Stag3 | 420 | 0.605 | 0.1789 | Yes |
| 5 | Skil | 673 | 0.513 | 0.1938 | Yes |
| 6 | Snn | 783 | 0.489 | 0.2187 | Yes |
| 7 | Ypel1 | 893 | 0.468 | 0.2421 | Yes |
| 8 | Tent5c | 953 | 0.460 | 0.2690 | Yes |
| 9 | Cpeb3 | 1411 | 0.386 | 0.2586 | Yes |
| 10 | Clstn3 | 1655 | 0.346 | 0.2628 | Yes |
| 11 | Tgfb2 | 2032 | 0.298 | 0.2529 | Yes |
| 12 | Selenop | 2179 | 0.283 | 0.2606 | Yes |
| 13 | Ccdc106 | 2339 | 0.268 | 0.2662 | Yes |
| 14 | Prodh | 2416 | 0.265 | 0.2783 | Yes |
| 15 | Fggy | 2578 | 0.248 | 0.2824 | Yes |
| 16 | Mx2 | 2616 | 0.245 | 0.2963 | Yes |
| 17 | Itgb1bp2 | 2707 | 0.235 | 0.3052 | Yes |
| 18 | Myo15a | 3226 | 0.196 | 0.2768 | No |
| 19 | Klk8 | 3292 | 0.191 | 0.2848 | No |
| 20 | Pdk2 | 3748 | 0.153 | 0.2585 | No |
| 21 | Rsad2 | 4207 | 0.118 | 0.2297 | No |
| 22 | Egf | 4746 | 0.081 | 0.1918 | No |
| 23 | Entpd7 | 5087 | 0.057 | 0.1683 | No |
| 24 | Lfng | 5278 | 0.045 | 0.1560 | No |
| 25 | Kmt2d | 5539 | 0.029 | 0.1370 | No |
| 26 | Zbtb16 | 5555 | 0.027 | 0.1377 | No |
| 27 | Brdt | 5617 | 0.023 | 0.1344 | No |
| 28 | Cyp39a1 | 5877 | 0.007 | 0.1140 | No |
| 29 | Tg | 5971 | 0.002 | 0.1066 | No |
| 30 | Grid2 | 6040 | 0.000 | 0.1011 | No |
| 31 | Prkn | 6189 | -0.006 | 0.0895 | No |
| 32 | Cdkal1 | 6201 | -0.007 | 0.0891 | No |
| 33 | Msh5 | 6400 | -0.020 | 0.0745 | No |
| 34 | Slc29a3 | 6702 | -0.038 | 0.0528 | No |
| 35 | Mthfr | 6754 | -0.041 | 0.0515 | No |
| 36 | Nr6a1 | 6801 | -0.044 | 0.0508 | No |
| 37 | Asb7 | 7176 | -0.067 | 0.0252 | No |
| 38 | Slc25a23 | 7298 | -0.074 | 0.0205 | No |
| 39 | Ptprj | 7816 | -0.109 | -0.0137 | No |
| 40 | Gpr19 | 7837 | -0.111 | -0.0077 | No |
| 41 | Dtnb | 8264 | -0.138 | -0.0326 | No |
| 42 | Thrb | 8376 | -0.146 | -0.0315 | No |
| 43 | Vps50 | 8539 | -0.157 | -0.0338 | No |
| 44 | Sgk1 | 8619 | -0.164 | -0.0289 | No |
| 45 | Arpp21 | 8625 | -0.165 | -0.0179 | No |
| 46 | Mfsd6 | 8666 | -0.169 | -0.0096 | No |
| 47 | Btg2 | 8796 | -0.177 | -0.0078 | No |
| 48 | Ybx2 | 9338 | -0.217 | -0.0366 | No |
| 49 | Slc16a7 | 9449 | -0.225 | -0.0300 | No |
| 50 | Tcf7l1 | 9489 | -0.226 | -0.0176 | No |
| 51 | Idua | 9673 | -0.240 | -0.0159 | No |
| 52 | Copz2 | 10350 | -0.297 | -0.0500 | No |
| 53 | Sphk2 | 10905 | -0.356 | -0.0703 | No |
| 54 | Gtf3c5 | 11022 | -0.372 | -0.0540 | No |
| 55 | Nr4a2 | 11159 | -0.391 | -0.0381 | No |
| 56 | Mast3 | 11293 | -0.411 | -0.0206 | No |
| 57 | Kcnd1 | 11663 | -0.474 | -0.0177 | No |
| 58 | Bard1 | 11886 | -0.528 | 0.0006 | No |
| 59 | Plag1 | 12170 | -0.641 | 0.0219 | No |