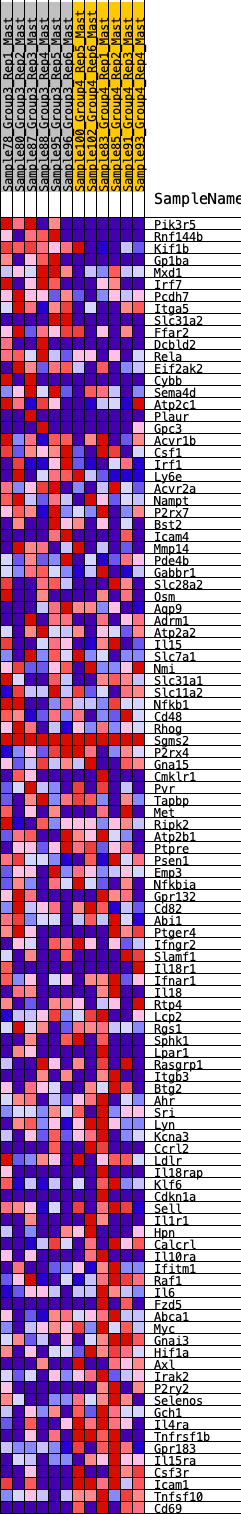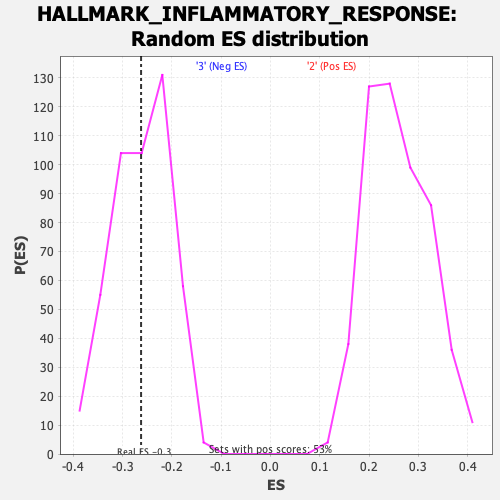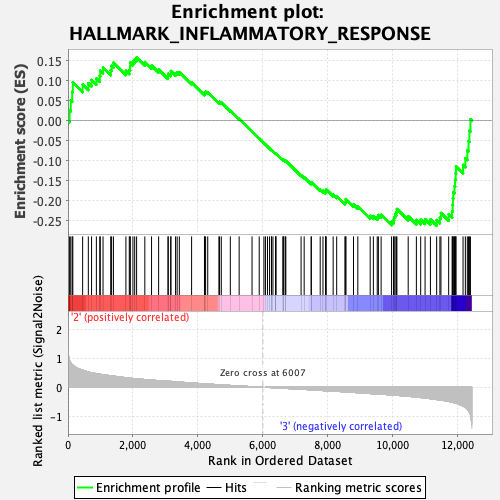
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.26247087 |
| Normalized Enrichment Score (NES) | -1.0086302 |
| Nominal p-value | 0.4713376 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pik3r5 | 49 | 0.918 | 0.0259 | No |
| 2 | Rnf144b | 85 | 0.848 | 0.0507 | No |
| 3 | Kif1b | 130 | 0.780 | 0.0726 | No |
| 4 | Gp1ba | 149 | 0.759 | 0.0958 | No |
| 5 | Mxd1 | 451 | 0.591 | 0.0907 | No |
| 6 | Irf7 | 626 | 0.528 | 0.0938 | No |
| 7 | Pcdh7 | 726 | 0.501 | 0.1021 | No |
| 8 | Itga5 | 874 | 0.471 | 0.1055 | No |
| 9 | Slc31a2 | 979 | 0.454 | 0.1118 | No |
| 10 | Ffar2 | 994 | 0.451 | 0.1254 | No |
| 11 | Dcbld2 | 1079 | 0.429 | 0.1326 | No |
| 12 | Rela | 1316 | 0.398 | 0.1264 | No |
| 13 | Eif2ak2 | 1343 | 0.394 | 0.1371 | No |
| 14 | Cybb | 1400 | 0.386 | 0.1451 | No |
| 15 | Sema4d | 1784 | 0.330 | 0.1248 | No |
| 16 | Atp2c1 | 1888 | 0.317 | 0.1268 | No |
| 17 | Plaur | 1913 | 0.313 | 0.1351 | No |
| 18 | Gpc3 | 1916 | 0.313 | 0.1451 | No |
| 19 | Acvr1b | 2000 | 0.302 | 0.1482 | No |
| 20 | Csf1 | 2055 | 0.297 | 0.1535 | No |
| 21 | Irf1 | 2114 | 0.291 | 0.1583 | No |
| 22 | Ly6e | 2367 | 0.267 | 0.1466 | No |
| 23 | Acvr2a | 2574 | 0.249 | 0.1380 | No |
| 24 | Nampt | 2792 | 0.228 | 0.1278 | No |
| 25 | P2rx7 | 3081 | 0.209 | 0.1113 | No |
| 26 | Bst2 | 3088 | 0.208 | 0.1176 | No |
| 27 | Icam4 | 3165 | 0.202 | 0.1180 | No |
| 28 | Mmp14 | 3171 | 0.202 | 0.1242 | No |
| 29 | Pde4b | 3314 | 0.189 | 0.1188 | No |
| 30 | Gabbr1 | 3363 | 0.185 | 0.1210 | No |
| 31 | Slc28a2 | 3430 | 0.180 | 0.1215 | No |
| 32 | Osm | 3808 | 0.149 | 0.0957 | No |
| 33 | Aqp9 | 4204 | 0.119 | 0.0676 | No |
| 34 | Adrm1 | 4211 | 0.118 | 0.0709 | No |
| 35 | Atp2a2 | 4239 | 0.116 | 0.0725 | No |
| 36 | Il15 | 4305 | 0.111 | 0.0709 | No |
| 37 | Slc7a1 | 4649 | 0.088 | 0.0459 | No |
| 38 | Nmi | 4670 | 0.086 | 0.0471 | No |
| 39 | Slc31a1 | 4723 | 0.082 | 0.0455 | No |
| 40 | Slc11a2 | 5001 | 0.063 | 0.0251 | No |
| 41 | Nfkb1 | 5273 | 0.046 | 0.0047 | No |
| 42 | Cd48 | 5670 | 0.020 | -0.0268 | No |
| 43 | Rhog | 5891 | 0.006 | -0.0444 | No |
| 44 | Sgms2 | 6036 | 0.000 | -0.0561 | No |
| 45 | P2rx4 | 6083 | -0.000 | -0.0598 | No |
| 46 | Gna15 | 6146 | -0.003 | -0.0647 | No |
| 47 | Cmklr1 | 6210 | -0.007 | -0.0696 | No |
| 48 | Pvr | 6269 | -0.010 | -0.0740 | No |
| 49 | Tapbp | 6270 | -0.010 | -0.0736 | No |
| 50 | Met | 6314 | -0.014 | -0.0767 | No |
| 51 | Ripk2 | 6397 | -0.019 | -0.0827 | No |
| 52 | Atp2b1 | 6398 | -0.019 | -0.0821 | No |
| 53 | Ptpre | 6407 | -0.020 | -0.0821 | No |
| 54 | Psen1 | 6623 | -0.033 | -0.0984 | No |
| 55 | Emp3 | 6627 | -0.033 | -0.0976 | No |
| 56 | Nfkbia | 6649 | -0.035 | -0.0982 | No |
| 57 | Gpr132 | 6696 | -0.038 | -0.1007 | No |
| 58 | Cd82 | 6711 | -0.039 | -0.1005 | No |
| 59 | Abi1 | 7181 | -0.067 | -0.1364 | No |
| 60 | Ptger4 | 7276 | -0.073 | -0.1416 | No |
| 61 | Ifngr2 | 7491 | -0.086 | -0.1562 | No |
| 62 | Slamf1 | 7497 | -0.086 | -0.1538 | No |
| 63 | Il18r1 | 7770 | -0.105 | -0.1724 | No |
| 64 | Ifnar1 | 7855 | -0.112 | -0.1755 | No |
| 65 | Il18 | 7935 | -0.117 | -0.1781 | No |
| 66 | Rtp4 | 7939 | -0.117 | -0.1746 | No |
| 67 | Lcp2 | 7955 | -0.118 | -0.1719 | No |
| 68 | Rgs1 | 8169 | -0.132 | -0.1849 | No |
| 69 | Sphk1 | 8278 | -0.139 | -0.1891 | No |
| 70 | Lpar1 | 8531 | -0.156 | -0.2045 | No |
| 71 | Rasgrp1 | 8558 | -0.159 | -0.2014 | No |
| 72 | Itgb3 | 8559 | -0.160 | -0.1962 | No |
| 73 | Btg2 | 8796 | -0.177 | -0.2096 | No |
| 74 | Ahr | 8929 | -0.187 | -0.2142 | No |
| 75 | Sri | 9310 | -0.215 | -0.2380 | No |
| 76 | Lyn | 9408 | -0.222 | -0.2386 | No |
| 77 | Kcna3 | 9527 | -0.227 | -0.2408 | No |
| 78 | Ccrl2 | 9563 | -0.230 | -0.2361 | No |
| 79 | Ldlr | 9647 | -0.237 | -0.2351 | No |
| 80 | Il18rap | 9969 | -0.264 | -0.2525 | Yes |
| 81 | Klf6 | 10029 | -0.269 | -0.2485 | Yes |
| 82 | Cdkn1a | 10046 | -0.270 | -0.2410 | Yes |
| 83 | Sell | 10077 | -0.273 | -0.2346 | Yes |
| 84 | Il1r1 | 10116 | -0.276 | -0.2287 | Yes |
| 85 | Hpn | 10133 | -0.278 | -0.2209 | Yes |
| 86 | Calcrl | 10481 | -0.309 | -0.2390 | Yes |
| 87 | Il10ra | 10733 | -0.337 | -0.2484 | Yes |
| 88 | Ifitm1 | 10866 | -0.352 | -0.2476 | Yes |
| 89 | Raf1 | 11001 | -0.369 | -0.2465 | Yes |
| 90 | Il6 | 11166 | -0.392 | -0.2470 | Yes |
| 91 | Fzd5 | 11358 | -0.422 | -0.2487 | Yes |
| 92 | Abca1 | 11458 | -0.433 | -0.2427 | Yes |
| 93 | Myc | 11490 | -0.438 | -0.2309 | Yes |
| 94 | Gnai3 | 11728 | -0.485 | -0.2343 | Yes |
| 95 | Hif1a | 11834 | -0.506 | -0.2263 | Yes |
| 96 | Axl | 11851 | -0.513 | -0.2109 | Yes |
| 97 | Irak2 | 11859 | -0.515 | -0.1947 | Yes |
| 98 | P2ry2 | 11871 | -0.521 | -0.1786 | Yes |
| 99 | Selenos | 11908 | -0.538 | -0.1640 | Yes |
| 100 | Gch1 | 11927 | -0.543 | -0.1478 | Yes |
| 101 | Il4ra | 11944 | -0.547 | -0.1313 | Yes |
| 102 | Tnfrsf1b | 11954 | -0.551 | -0.1140 | Yes |
| 103 | Gpr183 | 12171 | -0.641 | -0.1107 | Yes |
| 104 | Il15ra | 12242 | -0.692 | -0.0938 | Yes |
| 105 | Csf3r | 12303 | -0.753 | -0.0741 | Yes |
| 106 | Icam1 | 12340 | -0.803 | -0.0509 | Yes |
| 107 | Tnfsf10 | 12367 | -0.861 | -0.0250 | Yes |
| 108 | Cd69 | 12399 | -0.948 | 0.0034 | Yes |