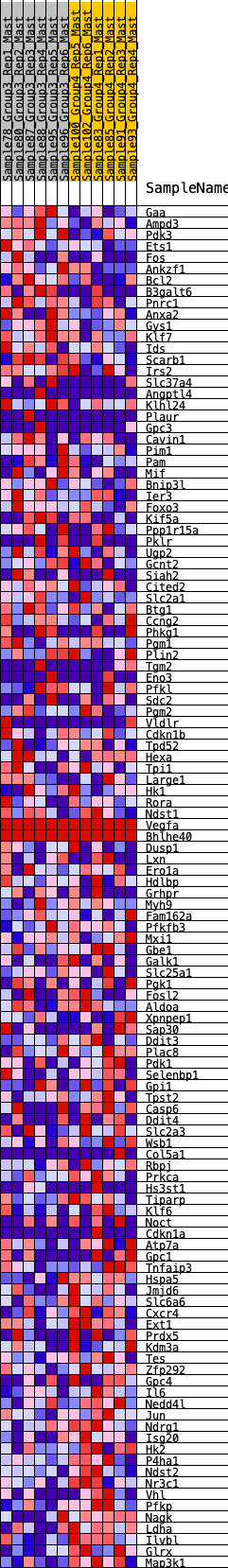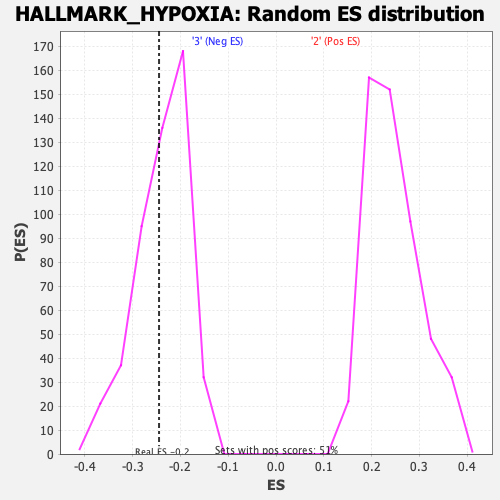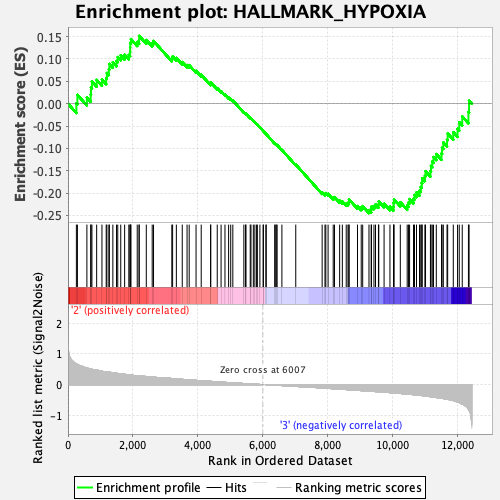
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | -0.2448725 |
| Normalized Enrichment Score (NES) | -1.0290002 |
| Nominal p-value | 0.38900203 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gaa | 258 | 0.677 | 0.0009 | No |
| 2 | Ampd3 | 290 | 0.661 | 0.0197 | No |
| 3 | Pdk3 | 582 | 0.543 | 0.0136 | No |
| 4 | Ets1 | 700 | 0.507 | 0.0204 | No |
| 5 | Fos | 708 | 0.504 | 0.0361 | No |
| 6 | Ankzf1 | 737 | 0.498 | 0.0499 | No |
| 7 | Bcl2 | 883 | 0.469 | 0.0532 | No |
| 8 | B3galt6 | 1046 | 0.436 | 0.0541 | No |
| 9 | Pnrc1 | 1177 | 0.416 | 0.0570 | No |
| 10 | Anxa2 | 1198 | 0.415 | 0.0687 | No |
| 11 | Gys1 | 1260 | 0.407 | 0.0769 | No |
| 12 | Klf7 | 1274 | 0.405 | 0.0889 | No |
| 13 | Ids | 1382 | 0.388 | 0.0927 | No |
| 14 | Scarb1 | 1494 | 0.369 | 0.0956 | No |
| 15 | Irs2 | 1533 | 0.364 | 0.1043 | No |
| 16 | Slc37a4 | 1625 | 0.350 | 0.1082 | No |
| 17 | Angptl4 | 1745 | 0.336 | 0.1093 | No |
| 18 | Klhl24 | 1874 | 0.318 | 0.1092 | No |
| 19 | Plaur | 1913 | 0.313 | 0.1162 | No |
| 20 | Gpc3 | 1916 | 0.313 | 0.1261 | No |
| 21 | Cavin1 | 1918 | 0.313 | 0.1362 | No |
| 22 | Pim1 | 1940 | 0.311 | 0.1445 | No |
| 23 | Pam | 2133 | 0.288 | 0.1382 | No |
| 24 | Mif | 2191 | 0.282 | 0.1426 | No |
| 25 | Bnip3l | 2192 | 0.282 | 0.1517 | No |
| 26 | Ier3 | 2412 | 0.265 | 0.1425 | No |
| 27 | Foxo3 | 2594 | 0.247 | 0.1358 | No |
| 28 | Kif5a | 2635 | 0.243 | 0.1404 | No |
| 29 | Ppp1r15a | 3199 | 0.199 | 0.1011 | No |
| 30 | Pklr | 3223 | 0.197 | 0.1056 | No |
| 31 | Ugp2 | 3338 | 0.188 | 0.1024 | No |
| 32 | Gcnt2 | 3520 | 0.171 | 0.0932 | No |
| 33 | Siah2 | 3668 | 0.159 | 0.0864 | No |
| 34 | Cited2 | 3734 | 0.154 | 0.0861 | No |
| 35 | Slc2a1 | 3945 | 0.138 | 0.0735 | No |
| 36 | Btg1 | 4104 | 0.126 | 0.0647 | No |
| 37 | Ccng2 | 4395 | 0.106 | 0.0446 | No |
| 38 | Phkg1 | 4396 | 0.106 | 0.0481 | No |
| 39 | Pgm1 | 4598 | 0.091 | 0.0347 | No |
| 40 | Plin2 | 4716 | 0.082 | 0.0278 | No |
| 41 | Tgm2 | 4836 | 0.074 | 0.0206 | No |
| 42 | Eno3 | 4948 | 0.067 | 0.0137 | No |
| 43 | Pfkl | 5013 | 0.062 | 0.0105 | No |
| 44 | Sdc2 | 5078 | 0.058 | 0.0072 | No |
| 45 | Pgm2 | 5418 | 0.036 | -0.0191 | No |
| 46 | Vldlr | 5470 | 0.033 | -0.0222 | No |
| 47 | Cdkn1b | 5483 | 0.033 | -0.0221 | No |
| 48 | Tpd52 | 5614 | 0.024 | -0.0319 | No |
| 49 | Hexa | 5637 | 0.022 | -0.0330 | No |
| 50 | Tpi1 | 5710 | 0.018 | -0.0382 | No |
| 51 | Large1 | 5757 | 0.016 | -0.0414 | No |
| 52 | Hk1 | 5810 | 0.013 | -0.0453 | No |
| 53 | Rora | 5834 | 0.011 | -0.0468 | No |
| 54 | Ndst1 | 5912 | 0.005 | -0.0528 | No |
| 55 | Vegfa | 6012 | 0.000 | -0.0609 | No |
| 56 | Bhlhe40 | 6020 | 0.000 | -0.0614 | No |
| 57 | Dusp1 | 6098 | -0.001 | -0.0676 | No |
| 58 | Lxn | 6106 | -0.002 | -0.0682 | No |
| 59 | Ero1a | 6367 | -0.018 | -0.0887 | No |
| 60 | Hdlbp | 6401 | -0.020 | -0.0907 | No |
| 61 | Grhpr | 6416 | -0.021 | -0.0912 | No |
| 62 | Myh9 | 6443 | -0.023 | -0.0926 | No |
| 63 | Fam162a | 6590 | -0.030 | -0.1034 | No |
| 64 | Pfkfb3 | 7017 | -0.057 | -0.1362 | No |
| 65 | Mxi1 | 7830 | -0.110 | -0.1985 | No |
| 66 | Gbe1 | 7917 | -0.116 | -0.2018 | No |
| 67 | Galk1 | 7943 | -0.117 | -0.2000 | No |
| 68 | Slc25a1 | 8013 | -0.122 | -0.2017 | No |
| 69 | Pgk1 | 8171 | -0.132 | -0.2102 | No |
| 70 | Fosl2 | 8210 | -0.135 | -0.2089 | No |
| 71 | Aldoa | 8369 | -0.146 | -0.2170 | No |
| 72 | Xpnpep1 | 8452 | -0.150 | -0.2188 | No |
| 73 | Sap30 | 8572 | -0.160 | -0.2233 | No |
| 74 | Ddit3 | 8626 | -0.165 | -0.2223 | No |
| 75 | Plac8 | 8655 | -0.167 | -0.2192 | No |
| 76 | Pdk1 | 8659 | -0.168 | -0.2140 | No |
| 77 | Selenbp1 | 8919 | -0.186 | -0.2290 | No |
| 78 | Gpi1 | 9034 | -0.194 | -0.2320 | No |
| 79 | Tpst2 | 9079 | -0.198 | -0.2292 | No |
| 80 | Casp6 | 9273 | -0.212 | -0.2380 | Yes |
| 81 | Ddit4 | 9340 | -0.217 | -0.2364 | Yes |
| 82 | Slc2a3 | 9350 | -0.217 | -0.2301 | Yes |
| 83 | Wsb1 | 9426 | -0.223 | -0.2290 | Yes |
| 84 | Col5a1 | 9474 | -0.226 | -0.2255 | Yes |
| 85 | Rbpj | 9565 | -0.230 | -0.2254 | Yes |
| 86 | Prkca | 9575 | -0.231 | -0.2187 | Yes |
| 87 | Hs3st1 | 9738 | -0.245 | -0.2239 | Yes |
| 88 | Tiparp | 9919 | -0.260 | -0.2302 | Yes |
| 89 | Klf6 | 10029 | -0.269 | -0.2303 | Yes |
| 90 | Noct | 10033 | -0.270 | -0.2219 | Yes |
| 91 | Cdkn1a | 10046 | -0.270 | -0.2141 | Yes |
| 92 | Atp7a | 10238 | -0.286 | -0.2204 | Yes |
| 93 | Gpc1 | 10450 | -0.305 | -0.2277 | Yes |
| 94 | Tnfaip3 | 10492 | -0.310 | -0.2210 | Yes |
| 95 | Hspa5 | 10524 | -0.313 | -0.2135 | Yes |
| 96 | Jmjd6 | 10645 | -0.328 | -0.2126 | Yes |
| 97 | Slc6a6 | 10675 | -0.331 | -0.2043 | Yes |
| 98 | Cxcr4 | 10737 | -0.338 | -0.1984 | Yes |
| 99 | Ext1 | 10830 | -0.349 | -0.1946 | Yes |
| 100 | Prdx5 | 10869 | -0.353 | -0.1863 | Yes |
| 101 | Kdm3a | 10898 | -0.355 | -0.1771 | Yes |
| 102 | Tes | 10913 | -0.357 | -0.1667 | Yes |
| 103 | Zfp292 | 10996 | -0.368 | -0.1615 | Yes |
| 104 | Gpc4 | 11015 | -0.371 | -0.1510 | Yes |
| 105 | Il6 | 11166 | -0.392 | -0.1506 | Yes |
| 106 | Nedd4l | 11183 | -0.395 | -0.1391 | Yes |
| 107 | Jun | 11222 | -0.399 | -0.1293 | Yes |
| 108 | Ndrg1 | 11261 | -0.405 | -0.1194 | Yes |
| 109 | Isg20 | 11347 | -0.421 | -0.1127 | Yes |
| 110 | Hk2 | 11507 | -0.442 | -0.1113 | Yes |
| 111 | P4ha1 | 11525 | -0.446 | -0.0984 | Yes |
| 112 | Ndst2 | 11563 | -0.453 | -0.0868 | Yes |
| 113 | Nr3c1 | 11678 | -0.476 | -0.0807 | Yes |
| 114 | Vhl | 11697 | -0.480 | -0.0666 | Yes |
| 115 | Pfkp | 11870 | -0.521 | -0.0638 | Yes |
| 116 | Nagk | 12002 | -0.566 | -0.0562 | Yes |
| 117 | Ldha | 12054 | -0.585 | -0.0415 | Yes |
| 118 | Ilvbl | 12146 | -0.631 | -0.0285 | Yes |
| 119 | Glrx | 12337 | -0.792 | -0.0184 | Yes |
| 120 | Map3k1 | 12357 | -0.830 | 0.0068 | Yes |