Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
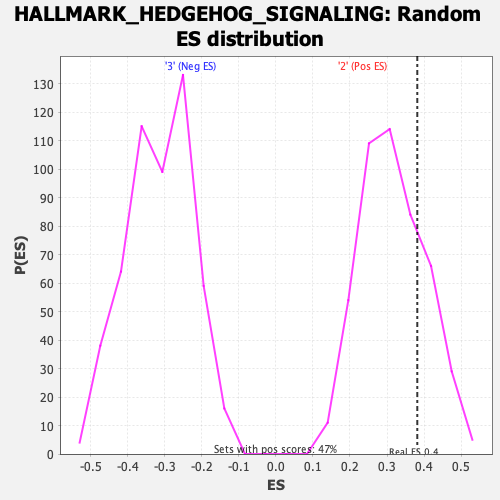
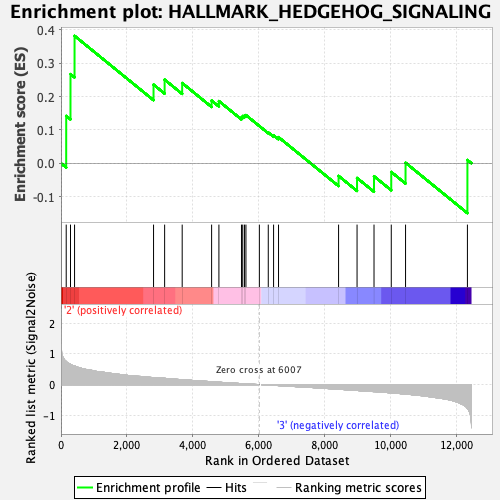
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.38167506 |
| Normalized Enrichment Score (NES) | 1.2034906 |
| Nominal p-value | 0.23516949 |
| FDR q-value | 0.5667509 |
| FWER p-Value | 0.929 |

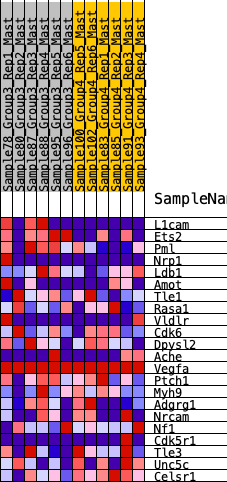
| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | L1cam | 158 | 0.754 | 0.1418 | Yes |
| 2 | Ets2 | 288 | 0.662 | 0.2671 | Yes |
| 3 | Pml | 411 | 0.607 | 0.3817 | Yes |
| 4 | Nrp1 | 2805 | 0.227 | 0.2356 | No |
| 5 | Ldb1 | 3141 | 0.204 | 0.2505 | No |
| 6 | Amot | 3673 | 0.159 | 0.2404 | No |
| 7 | Tle1 | 4565 | 0.094 | 0.1879 | No |
| 8 | Rasa1 | 4786 | 0.078 | 0.1862 | No |
| 9 | Vldlr | 5470 | 0.033 | 0.1381 | No |
| 10 | Cdk6 | 5508 | 0.031 | 0.1415 | No |
| 11 | Dpysl2 | 5560 | 0.027 | 0.1429 | No |
| 12 | Ache | 5605 | 0.024 | 0.1443 | No |
| 13 | Vegfa | 6012 | 0.000 | 0.1116 | No |
| 14 | Ptch1 | 6282 | -0.011 | 0.0922 | No |
| 15 | Myh9 | 6443 | -0.023 | 0.0840 | No |
| 16 | Adgrg1 | 6593 | -0.031 | 0.0783 | No |
| 17 | Nrcam | 8413 | -0.148 | -0.0379 | No |
| 18 | Nf1 | 8973 | -0.190 | -0.0440 | No |
| 19 | Cdk5r1 | 9487 | -0.226 | -0.0389 | No |
| 20 | Tle3 | 10010 | -0.268 | -0.0260 | No |
| 21 | Unc5c | 10443 | -0.304 | 0.0016 | No |
| 22 | Celsr1 | 12318 | -0.777 | 0.0099 | No |