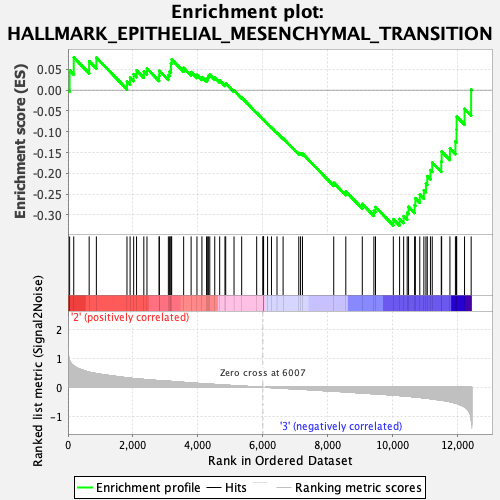
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
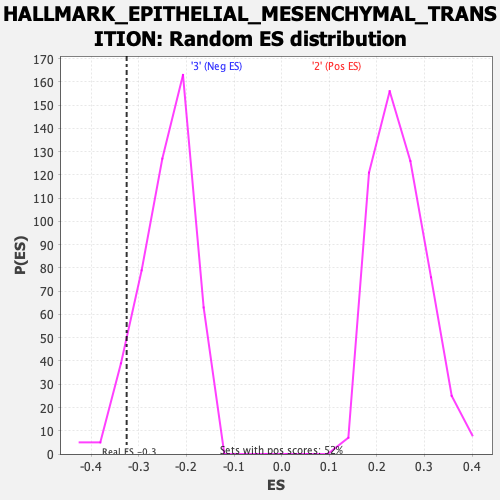
| Enrichment Score (ES) | -0.3256381 |
| Normalized Enrichment Score (NES) | -1.3449446 |
| Nominal p-value | 0.06652807 |
| FDR q-value | 0.891409 |
| FWER p-Value | 0.707 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sat1 | 50 | 0.914 | 0.0474 | No |
| 2 | Mmp2 | 177 | 0.734 | 0.0786 | No |
| 3 | Sgcb | 652 | 0.519 | 0.0695 | No |
| 4 | Itga5 | 874 | 0.471 | 0.0782 | No |
| 5 | Timp3 | 1817 | 0.325 | 0.0204 | No |
| 6 | Plaur | 1913 | 0.313 | 0.0303 | No |
| 7 | Copa | 2023 | 0.299 | 0.0384 | No |
| 8 | Notch2 | 2112 | 0.292 | 0.0477 | No |
| 9 | Dst | 2336 | 0.269 | 0.0448 | No |
| 10 | Itga2 | 2433 | 0.263 | 0.0519 | No |
| 11 | Fbn1 | 2806 | 0.227 | 0.0347 | No |
| 12 | Gja1 | 2815 | 0.227 | 0.0468 | No |
| 13 | Cd44 | 3092 | 0.208 | 0.0362 | No |
| 14 | Wipf1 | 3130 | 0.205 | 0.0448 | No |
| 15 | Mmp14 | 3171 | 0.202 | 0.0529 | No |
| 16 | Calu | 3172 | 0.202 | 0.0643 | No |
| 17 | Lamc1 | 3194 | 0.200 | 0.0739 | No |
| 18 | Fuca1 | 3563 | 0.168 | 0.0536 | No |
| 19 | Itgb1 | 3793 | 0.150 | 0.0435 | No |
| 20 | Capg | 3973 | 0.136 | 0.0367 | No |
| 21 | Cald1 | 4126 | 0.124 | 0.0315 | No |
| 22 | Tgfbr3 | 4269 | 0.113 | 0.0264 | No |
| 23 | Il15 | 4305 | 0.111 | 0.0298 | No |
| 24 | P3h1 | 4323 | 0.111 | 0.0346 | No |
| 25 | Lgals1 | 4368 | 0.108 | 0.0372 | No |
| 26 | Tpm4 | 4521 | 0.097 | 0.0303 | No |
| 27 | Rhob | 4673 | 0.085 | 0.0229 | No |
| 28 | Tgm2 | 4836 | 0.074 | 0.0140 | No |
| 29 | Cap2 | 4859 | 0.073 | 0.0163 | No |
| 30 | Bmp1 | 5115 | 0.056 | -0.0012 | No |
| 31 | Colgalt1 | 5351 | 0.040 | -0.0179 | No |
| 32 | Id2 | 5813 | 0.012 | -0.0545 | No |
| 33 | Vcan | 6010 | 0.000 | -0.0703 | No |
| 34 | Vegfa | 6012 | 0.000 | -0.0704 | No |
| 35 | Serpine2 | 6015 | 0.000 | -0.0705 | No |
| 36 | Thbs1 | 6026 | 0.000 | -0.0714 | No |
| 37 | Gem | 6149 | -0.003 | -0.0810 | No |
| 38 | Pvr | 6269 | -0.010 | -0.0901 | No |
| 39 | Glipr1 | 6439 | -0.023 | -0.1024 | No |
| 40 | Emp3 | 6627 | -0.033 | -0.1157 | No |
| 41 | Plod1 | 7110 | -0.064 | -0.1511 | No |
| 42 | Ecm2 | 7165 | -0.067 | -0.1517 | No |
| 43 | Tgfb1 | 7224 | -0.069 | -0.1525 | No |
| 44 | Plod2 | 8187 | -0.133 | -0.2227 | No |
| 45 | Itgb3 | 8559 | -0.160 | -0.2437 | No |
| 46 | Sntb1 | 9067 | -0.197 | -0.2736 | No |
| 47 | Igfbp4 | 9424 | -0.223 | -0.2898 | No |
| 48 | Col5a1 | 9474 | -0.226 | -0.2810 | No |
| 49 | Ntm | 10023 | -0.269 | -0.3101 | Yes |
| 50 | Vim | 10216 | -0.284 | -0.3096 | Yes |
| 51 | Tpm1 | 10341 | -0.296 | -0.3030 | Yes |
| 52 | Gpc1 | 10450 | -0.305 | -0.2945 | Yes |
| 53 | Tnfaip3 | 10492 | -0.310 | -0.2803 | Yes |
| 54 | Ecm1 | 10678 | -0.331 | -0.2766 | Yes |
| 55 | Itgav | 10702 | -0.334 | -0.2597 | Yes |
| 56 | Pmepa1 | 10838 | -0.350 | -0.2509 | Yes |
| 57 | Sfrp4 | 10964 | -0.364 | -0.2405 | Yes |
| 58 | Flna | 11028 | -0.372 | -0.2246 | Yes |
| 59 | Plod3 | 11069 | -0.379 | -0.2065 | Yes |
| 60 | Il6 | 11166 | -0.392 | -0.1922 | Yes |
| 61 | Jun | 11222 | -0.399 | -0.1741 | Yes |
| 62 | Pdgfrb | 11500 | -0.441 | -0.1717 | Yes |
| 63 | Fap | 11512 | -0.443 | -0.1477 | Yes |
| 64 | Vegfc | 11767 | -0.494 | -0.1404 | Yes |
| 65 | Qsox1 | 11936 | -0.545 | -0.1233 | Yes |
| 66 | Mcm7 | 11971 | -0.556 | -0.0947 | Yes |
| 67 | Ppib | 11974 | -0.557 | -0.0635 | Yes |
| 68 | Gadd45a | 12217 | -0.676 | -0.0450 | Yes |
| 69 | Basp1 | 12421 | -1.119 | 0.0016 | Yes |