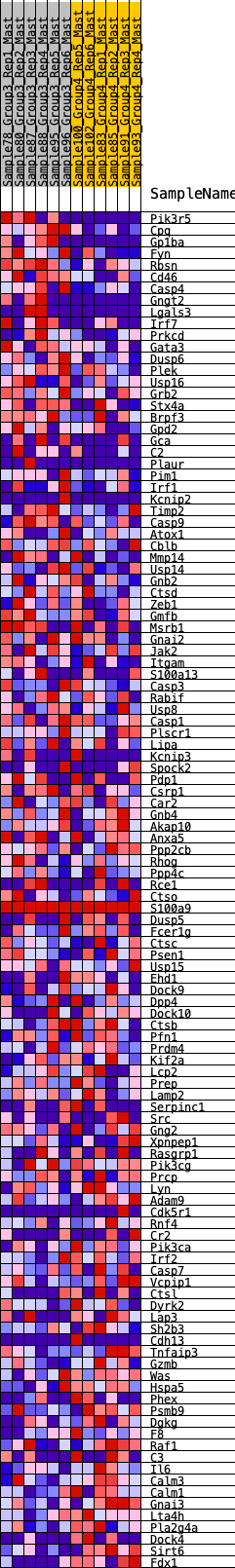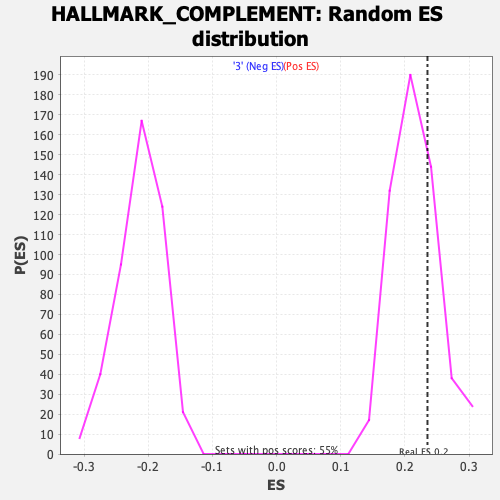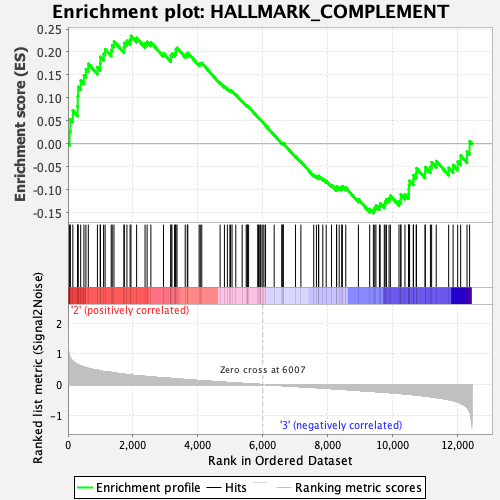
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.23507524 |
| Normalized Enrichment Score (NES) | 1.087438 |
| Nominal p-value | 0.26422018 |
| FDR q-value | 0.63485277 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pik3r5 | 49 | 0.918 | 0.0265 | Yes |
| 2 | Cpq | 73 | 0.863 | 0.0533 | Yes |
| 3 | Gp1ba | 149 | 0.759 | 0.0724 | Yes |
| 4 | Fyn | 297 | 0.653 | 0.0822 | Yes |
| 5 | Rbsn | 301 | 0.651 | 0.1036 | Yes |
| 6 | Cd46 | 313 | 0.645 | 0.1241 | Yes |
| 7 | Casp4 | 391 | 0.611 | 0.1382 | Yes |
| 8 | Gngt2 | 491 | 0.573 | 0.1492 | Yes |
| 9 | Lgals3 | 552 | 0.549 | 0.1625 | Yes |
| 10 | Irf7 | 626 | 0.528 | 0.1742 | Yes |
| 11 | Prkcd | 906 | 0.467 | 0.1670 | Yes |
| 12 | Gata3 | 993 | 0.452 | 0.1751 | Yes |
| 13 | Dusp6 | 999 | 0.450 | 0.1896 | Yes |
| 14 | Plek | 1097 | 0.426 | 0.1959 | Yes |
| 15 | Usp16 | 1143 | 0.420 | 0.2062 | Yes |
| 16 | Grb2 | 1331 | 0.395 | 0.2041 | Yes |
| 17 | Stx4a | 1362 | 0.392 | 0.2147 | Yes |
| 18 | Brpf3 | 1416 | 0.384 | 0.2232 | Yes |
| 19 | Gpd2 | 1722 | 0.339 | 0.2097 | Yes |
| 20 | Gca | 1736 | 0.337 | 0.2198 | Yes |
| 21 | C2 | 1815 | 0.325 | 0.2243 | Yes |
| 22 | Plaur | 1913 | 0.313 | 0.2269 | Yes |
| 23 | Pim1 | 1940 | 0.311 | 0.2351 | Yes |
| 24 | Irf1 | 2114 | 0.291 | 0.2307 | No |
| 25 | Kcnip2 | 2374 | 0.267 | 0.2186 | No |
| 26 | Timp2 | 2438 | 0.263 | 0.2222 | No |
| 27 | Casp9 | 2553 | 0.251 | 0.2213 | No |
| 28 | Atox1 | 2942 | 0.216 | 0.1970 | No |
| 29 | Cblb | 3167 | 0.202 | 0.1855 | No |
| 30 | Mmp14 | 3171 | 0.202 | 0.1920 | No |
| 31 | Usp14 | 3203 | 0.199 | 0.1961 | No |
| 32 | Gnb2 | 3283 | 0.192 | 0.1960 | No |
| 33 | Ctsd | 3319 | 0.189 | 0.1995 | No |
| 34 | Zeb1 | 3320 | 0.189 | 0.2057 | No |
| 35 | Gmfb | 3358 | 0.186 | 0.2089 | No |
| 36 | Msrb1 | 3611 | 0.164 | 0.1939 | No |
| 37 | Gnai2 | 3675 | 0.159 | 0.1941 | No |
| 38 | Jak2 | 3691 | 0.158 | 0.1981 | No |
| 39 | Itgam | 4040 | 0.130 | 0.1742 | No |
| 40 | S100a13 | 4076 | 0.128 | 0.1756 | No |
| 41 | Casp3 | 4122 | 0.125 | 0.1761 | No |
| 42 | Rabif | 4686 | 0.084 | 0.1332 | No |
| 43 | Usp8 | 4819 | 0.076 | 0.1250 | No |
| 44 | Casp1 | 4910 | 0.069 | 0.1200 | No |
| 45 | Plscr1 | 4987 | 0.065 | 0.1160 | No |
| 46 | Lipa | 5009 | 0.063 | 0.1164 | No |
| 47 | Kcnip3 | 5054 | 0.060 | 0.1148 | No |
| 48 | Spock2 | 5169 | 0.052 | 0.1073 | No |
| 49 | Pdp1 | 5366 | 0.039 | 0.0927 | No |
| 50 | Csrp1 | 5489 | 0.033 | 0.0839 | No |
| 51 | Car2 | 5520 | 0.030 | 0.0824 | No |
| 52 | Gnb4 | 5526 | 0.030 | 0.0830 | No |
| 53 | Akap10 | 5564 | 0.027 | 0.0809 | No |
| 54 | Anxa5 | 5845 | 0.011 | 0.0585 | No |
| 55 | Ppp2cb | 5872 | 0.008 | 0.0567 | No |
| 56 | Rhog | 5891 | 0.006 | 0.0554 | No |
| 57 | Ppp4c | 5938 | 0.004 | 0.0518 | No |
| 58 | Rce1 | 5943 | 0.004 | 0.0517 | No |
| 59 | Ctso | 6002 | 0.000 | 0.0470 | No |
| 60 | S100a9 | 6037 | 0.000 | 0.0442 | No |
| 61 | Dusp5 | 6086 | -0.001 | 0.0403 | No |
| 62 | Fcer1g | 6352 | -0.017 | 0.0194 | No |
| 63 | Ctsc | 6584 | -0.030 | 0.0016 | No |
| 64 | Psen1 | 6623 | -0.033 | -0.0004 | No |
| 65 | Usp15 | 6624 | -0.033 | 0.0007 | No |
| 66 | Ehd1 | 6626 | -0.033 | 0.0018 | No |
| 67 | Dock9 | 7010 | -0.057 | -0.0274 | No |
| 68 | Dpp4 | 7175 | -0.067 | -0.0385 | No |
| 69 | Dock10 | 7571 | -0.091 | -0.0675 | No |
| 70 | Ctsb | 7654 | -0.097 | -0.0710 | No |
| 71 | Pfn1 | 7720 | -0.102 | -0.0729 | No |
| 72 | Prdm4 | 7723 | -0.102 | -0.0696 | No |
| 73 | Kif2a | 7850 | -0.112 | -0.0761 | No |
| 74 | Lcp2 | 7955 | -0.118 | -0.0807 | No |
| 75 | Prep | 8119 | -0.129 | -0.0896 | No |
| 76 | Lamp2 | 8273 | -0.139 | -0.0974 | No |
| 77 | Serpinc1 | 8277 | -0.139 | -0.0930 | No |
| 78 | Src | 8355 | -0.145 | -0.0945 | No |
| 79 | Gng2 | 8432 | -0.149 | -0.0957 | No |
| 80 | Xpnpep1 | 8452 | -0.150 | -0.0922 | No |
| 81 | Rasgrp1 | 8558 | -0.159 | -0.0955 | No |
| 82 | Pik3cg | 8947 | -0.188 | -0.1207 | No |
| 83 | Prcp | 9295 | -0.214 | -0.1417 | No |
| 84 | Lyn | 9408 | -0.222 | -0.1434 | No |
| 85 | Adam9 | 9451 | -0.225 | -0.1394 | No |
| 86 | Cdk5r1 | 9487 | -0.226 | -0.1347 | No |
| 87 | Rnf4 | 9594 | -0.233 | -0.1355 | No |
| 88 | Cr2 | 9618 | -0.235 | -0.1296 | No |
| 89 | Pik3ca | 9744 | -0.245 | -0.1316 | No |
| 90 | Irf2 | 9771 | -0.247 | -0.1255 | No |
| 91 | Casp7 | 9810 | -0.251 | -0.1202 | No |
| 92 | Vcpip1 | 9891 | -0.258 | -0.1182 | No |
| 93 | Ctsl | 9932 | -0.261 | -0.1127 | No |
| 94 | Dyrk2 | 10198 | -0.283 | -0.1249 | No |
| 95 | Lap3 | 10253 | -0.288 | -0.1197 | No |
| 96 | Sh2b3 | 10255 | -0.288 | -0.1102 | No |
| 97 | Cdh13 | 10381 | -0.298 | -0.1104 | No |
| 98 | Tnfaip3 | 10492 | -0.310 | -0.1090 | No |
| 99 | Gzmb | 10496 | -0.311 | -0.0990 | No |
| 100 | Was | 10506 | -0.311 | -0.0894 | No |
| 101 | Hspa5 | 10524 | -0.313 | -0.0803 | No |
| 102 | Phex | 10639 | -0.328 | -0.0787 | No |
| 103 | Psmb9 | 10642 | -0.328 | -0.0680 | No |
| 104 | Dgkg | 10727 | -0.337 | -0.0636 | No |
| 105 | F8 | 10736 | -0.337 | -0.0530 | No |
| 106 | Raf1 | 11001 | -0.369 | -0.0622 | No |
| 107 | C3 | 11007 | -0.369 | -0.0503 | No |
| 108 | Il6 | 11166 | -0.392 | -0.0501 | No |
| 109 | Calm3 | 11203 | -0.396 | -0.0399 | No |
| 110 | Calm1 | 11345 | -0.421 | -0.0374 | No |
| 111 | Gnai3 | 11728 | -0.485 | -0.0522 | No |
| 112 | Lta4h | 11864 | -0.518 | -0.0460 | No |
| 113 | Pla2g4a | 12006 | -0.567 | -0.0386 | No |
| 114 | Dock4 | 12097 | -0.612 | -0.0256 | No |
| 115 | Sirt6 | 12293 | -0.737 | -0.0169 | No |
| 116 | Fdx1 | 12373 | -0.868 | 0.0055 | No |