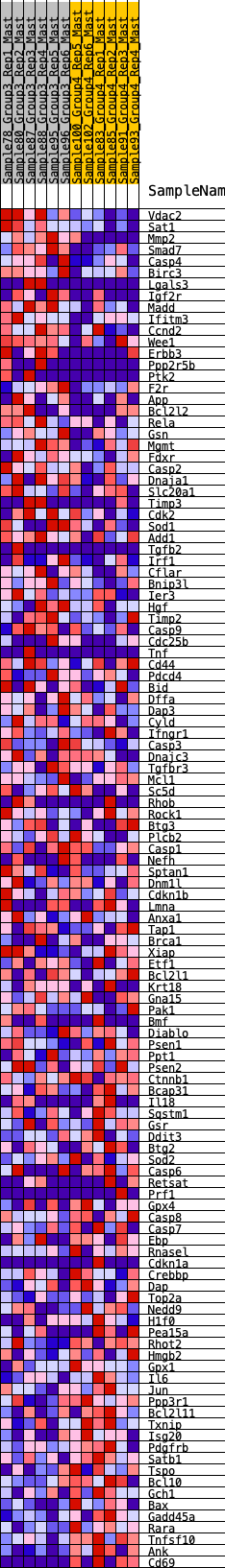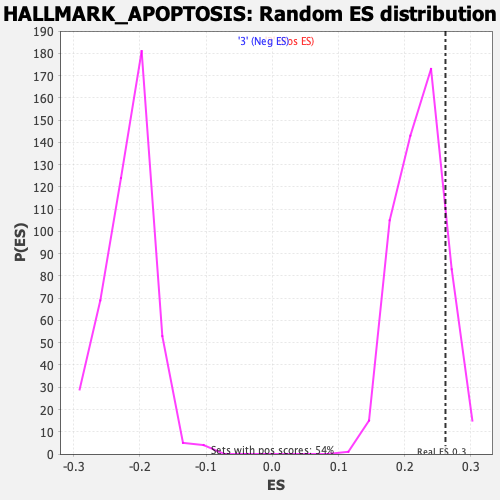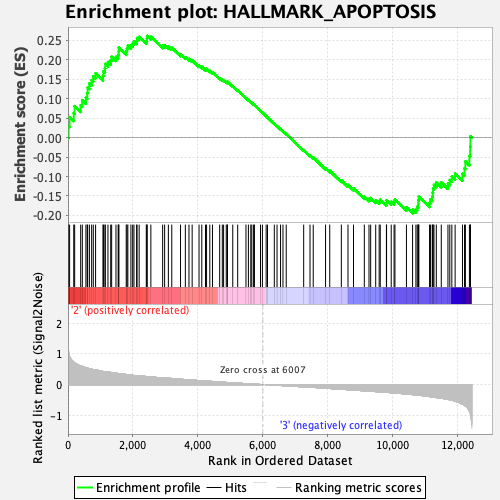
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.26155698 |
| Normalized Enrichment Score (NES) | 1.1755458 |
| Nominal p-value | 0.14579439 |
| FDR q-value | 0.5749681 |
| FWER p-Value | 0.961 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vdac2 | 19 | 1.090 | 0.0293 | Yes |
| 2 | Sat1 | 50 | 0.914 | 0.0527 | Yes |
| 3 | Mmp2 | 177 | 0.734 | 0.0632 | Yes |
| 4 | Smad7 | 205 | 0.710 | 0.0811 | Yes |
| 5 | Casp4 | 391 | 0.611 | 0.0833 | Yes |
| 6 | Birc3 | 441 | 0.593 | 0.0961 | Yes |
| 7 | Lgals3 | 552 | 0.549 | 0.1027 | Yes |
| 8 | Igf2r | 593 | 0.541 | 0.1148 | Yes |
| 9 | Madd | 605 | 0.537 | 0.1291 | Yes |
| 10 | Ifitm3 | 659 | 0.516 | 0.1393 | Yes |
| 11 | Ccnd2 | 729 | 0.500 | 0.1479 | Yes |
| 12 | Wee1 | 779 | 0.490 | 0.1578 | Yes |
| 13 | Erbb3 | 850 | 0.474 | 0.1655 | Yes |
| 14 | Ppp2r5b | 1078 | 0.429 | 0.1592 | Yes |
| 15 | Ptk2 | 1092 | 0.427 | 0.1702 | Yes |
| 16 | F2r | 1138 | 0.421 | 0.1785 | Yes |
| 17 | App | 1149 | 0.419 | 0.1895 | Yes |
| 18 | Bcl2l2 | 1231 | 0.409 | 0.1945 | Yes |
| 19 | Rela | 1316 | 0.398 | 0.1989 | Yes |
| 20 | Gsn | 1345 | 0.394 | 0.2078 | Yes |
| 21 | Mgmt | 1478 | 0.373 | 0.2076 | Yes |
| 22 | Fdxr | 1545 | 0.361 | 0.2125 | Yes |
| 23 | Casp2 | 1562 | 0.359 | 0.2213 | Yes |
| 24 | Dnaja1 | 1565 | 0.358 | 0.2313 | Yes |
| 25 | Slc20a1 | 1794 | 0.328 | 0.2221 | Yes |
| 26 | Timp3 | 1817 | 0.325 | 0.2295 | Yes |
| 27 | Cdk2 | 1844 | 0.321 | 0.2364 | Yes |
| 28 | Sod1 | 1938 | 0.311 | 0.2377 | Yes |
| 29 | Add1 | 1992 | 0.303 | 0.2420 | Yes |
| 30 | Tgfb2 | 2032 | 0.298 | 0.2472 | Yes |
| 31 | Irf1 | 2114 | 0.291 | 0.2489 | Yes |
| 32 | Cflar | 2127 | 0.290 | 0.2561 | Yes |
| 33 | Bnip3l | 2192 | 0.282 | 0.2589 | Yes |
| 34 | Ier3 | 2412 | 0.265 | 0.2486 | Yes |
| 35 | Hgf | 2431 | 0.263 | 0.2546 | Yes |
| 36 | Timp2 | 2438 | 0.263 | 0.2616 | Yes |
| 37 | Casp9 | 2553 | 0.251 | 0.2594 | No |
| 38 | Cdc25b | 2916 | 0.219 | 0.2362 | No |
| 39 | Tnf | 2977 | 0.214 | 0.2374 | No |
| 40 | Cd44 | 3092 | 0.208 | 0.2340 | No |
| 41 | Pdcd4 | 3197 | 0.200 | 0.2312 | No |
| 42 | Bid | 3467 | 0.175 | 0.2144 | No |
| 43 | Dffa | 3615 | 0.164 | 0.2071 | No |
| 44 | Dap3 | 3727 | 0.155 | 0.2024 | No |
| 45 | Cyld | 3825 | 0.147 | 0.1987 | No |
| 46 | Ifngr1 | 4037 | 0.131 | 0.1853 | No |
| 47 | Casp3 | 4122 | 0.125 | 0.1820 | No |
| 48 | Dnajc3 | 4236 | 0.116 | 0.1761 | No |
| 49 | Tgfbr3 | 4269 | 0.113 | 0.1767 | No |
| 50 | Mcl1 | 4369 | 0.108 | 0.1717 | No |
| 51 | Sc5d | 4451 | 0.102 | 0.1681 | No |
| 52 | Rhob | 4673 | 0.085 | 0.1525 | No |
| 53 | Rock1 | 4754 | 0.080 | 0.1483 | No |
| 54 | Btg3 | 4794 | 0.078 | 0.1474 | No |
| 55 | Plcb2 | 4876 | 0.071 | 0.1428 | No |
| 56 | Casp1 | 4910 | 0.069 | 0.1421 | No |
| 57 | Nefh | 4911 | 0.069 | 0.1440 | No |
| 58 | Sptan1 | 5076 | 0.059 | 0.1324 | No |
| 59 | Dnm1l | 5228 | 0.048 | 0.1215 | No |
| 60 | Cdkn1b | 5483 | 0.033 | 0.1018 | No |
| 61 | Lmna | 5558 | 0.027 | 0.0965 | No |
| 62 | Anxa1 | 5632 | 0.023 | 0.0913 | No |
| 63 | Tap1 | 5663 | 0.021 | 0.0894 | No |
| 64 | Brca1 | 5718 | 0.018 | 0.0855 | No |
| 65 | Xiap | 5744 | 0.016 | 0.0840 | No |
| 66 | Etf1 | 5931 | 0.005 | 0.0690 | No |
| 67 | Bcl2l1 | 5990 | 0.001 | 0.0643 | No |
| 68 | Krt18 | 6105 | -0.002 | 0.0551 | No |
| 69 | Gna15 | 6146 | -0.003 | 0.0520 | No |
| 70 | Pak1 | 6356 | -0.017 | 0.0355 | No |
| 71 | Bmf | 6440 | -0.023 | 0.0294 | No |
| 72 | Diablo | 6548 | -0.027 | 0.0215 | No |
| 73 | Psen1 | 6623 | -0.033 | 0.0164 | No |
| 74 | Ppt1 | 6723 | -0.040 | 0.0095 | No |
| 75 | Psen2 | 7261 | -0.072 | -0.0320 | No |
| 76 | Ctnnb1 | 7456 | -0.083 | -0.0454 | No |
| 77 | Bcap31 | 7555 | -0.090 | -0.0508 | No |
| 78 | Il18 | 7935 | -0.117 | -0.0783 | No |
| 79 | Sqstm1 | 8064 | -0.126 | -0.0851 | No |
| 80 | Gsr | 8421 | -0.149 | -0.1098 | No |
| 81 | Ddit3 | 8626 | -0.165 | -0.1217 | No |
| 82 | Btg2 | 8796 | -0.177 | -0.1304 | No |
| 83 | Sod2 | 9129 | -0.201 | -0.1517 | No |
| 84 | Casp6 | 9273 | -0.212 | -0.1573 | No |
| 85 | Retsat | 9325 | -0.216 | -0.1553 | No |
| 86 | Prf1 | 9480 | -0.226 | -0.1614 | No |
| 87 | Gpx4 | 9587 | -0.233 | -0.1634 | No |
| 88 | Casp8 | 9620 | -0.236 | -0.1594 | No |
| 89 | Casp7 | 9810 | -0.251 | -0.1676 | No |
| 90 | Ebp | 9815 | -0.251 | -0.1608 | No |
| 91 | Rnasel | 9958 | -0.264 | -0.1649 | No |
| 92 | Cdkn1a | 10046 | -0.270 | -0.1643 | No |
| 93 | Crebbp | 10073 | -0.272 | -0.1587 | No |
| 94 | Dap | 10427 | -0.303 | -0.1788 | No |
| 95 | Top2a | 10616 | -0.324 | -0.1849 | No |
| 96 | Nedd9 | 10719 | -0.336 | -0.1837 | No |
| 97 | H1f0 | 10760 | -0.341 | -0.1773 | No |
| 98 | Pea15a | 10794 | -0.344 | -0.1702 | No |
| 99 | Rhot2 | 10795 | -0.344 | -0.1605 | No |
| 100 | Hmgb2 | 10810 | -0.347 | -0.1518 | No |
| 101 | Gpx1 | 11139 | -0.389 | -0.1674 | No |
| 102 | Il6 | 11166 | -0.392 | -0.1585 | No |
| 103 | Jun | 11222 | -0.399 | -0.1516 | No |
| 104 | Ppp3r1 | 11237 | -0.402 | -0.1414 | No |
| 105 | Bcl2l11 | 11247 | -0.403 | -0.1307 | No |
| 106 | Txnip | 11275 | -0.407 | -0.1214 | No |
| 107 | Isg20 | 11347 | -0.421 | -0.1153 | No |
| 108 | Pdgfrb | 11500 | -0.441 | -0.1151 | No |
| 109 | Satb1 | 11706 | -0.482 | -0.1182 | No |
| 110 | Tspo | 11763 | -0.493 | -0.1088 | No |
| 111 | Bcl10 | 11826 | -0.503 | -0.0996 | No |
| 112 | Gch1 | 11927 | -0.543 | -0.0924 | No |
| 113 | Bax | 12153 | -0.633 | -0.0927 | No |
| 114 | Gadd45a | 12217 | -0.676 | -0.0787 | No |
| 115 | Rara | 12243 | -0.692 | -0.0612 | No |
| 116 | Tnfsf10 | 12367 | -0.861 | -0.0468 | No |
| 117 | Ank | 12392 | -0.916 | -0.0229 | No |
| 118 | Cd69 | 12399 | -0.948 | 0.0034 | No |