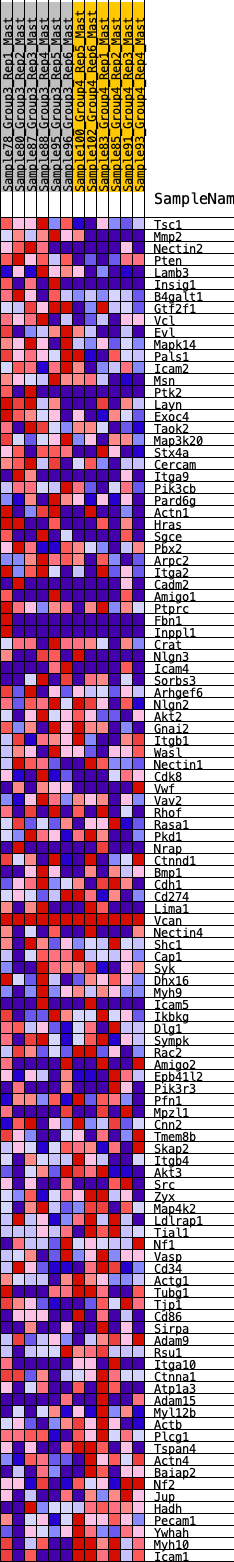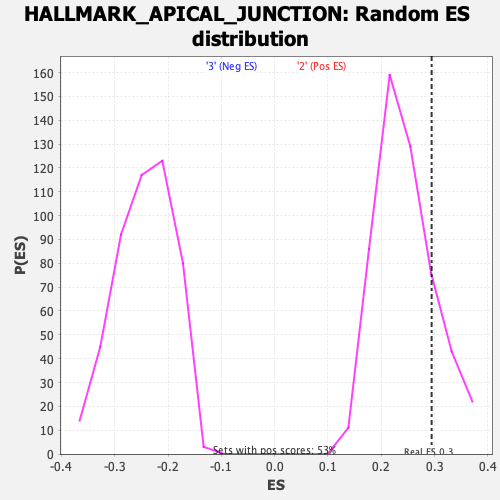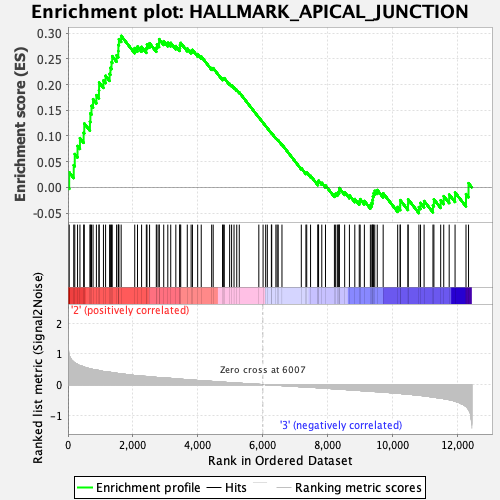
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group3_versus_Group4.Mast_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.2945398 |
| Normalized Enrichment Score (NES) | 1.2083473 |
| Nominal p-value | 0.17680608 |
| FDR q-value | 0.66431653 |
| FWER p-Value | 0.924 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tsc1 | 37 | 0.947 | 0.0294 | Yes |
| 2 | Mmp2 | 177 | 0.734 | 0.0432 | Yes |
| 3 | Nectin2 | 209 | 0.709 | 0.0649 | Yes |
| 4 | Pten | 294 | 0.656 | 0.0806 | Yes |
| 5 | Lamb3 | 368 | 0.619 | 0.0958 | Yes |
| 6 | Insig1 | 482 | 0.577 | 0.1064 | Yes |
| 7 | B4galt1 | 502 | 0.567 | 0.1242 | Yes |
| 8 | Gtf2f1 | 676 | 0.512 | 0.1277 | Yes |
| 9 | Vcl | 690 | 0.509 | 0.1441 | Yes |
| 10 | Evl | 724 | 0.501 | 0.1585 | Yes |
| 11 | Mapk14 | 772 | 0.491 | 0.1715 | Yes |
| 12 | Pals1 | 875 | 0.470 | 0.1793 | Yes |
| 13 | Icam2 | 954 | 0.459 | 0.1887 | Yes |
| 14 | Msn | 959 | 0.458 | 0.2040 | Yes |
| 15 | Ptk2 | 1092 | 0.427 | 0.2079 | Yes |
| 16 | Layn | 1158 | 0.418 | 0.2170 | Yes |
| 17 | Exoc4 | 1282 | 0.404 | 0.2208 | Yes |
| 18 | Taok2 | 1307 | 0.399 | 0.2325 | Yes |
| 19 | Map3k20 | 1341 | 0.394 | 0.2433 | Yes |
| 20 | Stx4a | 1362 | 0.392 | 0.2551 | Yes |
| 21 | Cercam | 1497 | 0.369 | 0.2569 | Yes |
| 22 | Itga9 | 1549 | 0.361 | 0.2651 | Yes |
| 23 | Pik3cb | 1553 | 0.360 | 0.2771 | Yes |
| 24 | Pard6g | 1569 | 0.358 | 0.2881 | Yes |
| 25 | Actn1 | 1638 | 0.348 | 0.2945 | Yes |
| 26 | Hras | 2056 | 0.297 | 0.2709 | No |
| 27 | Sgce | 2141 | 0.287 | 0.2739 | No |
| 28 | Pbx2 | 2267 | 0.275 | 0.2731 | No |
| 29 | Arpc2 | 2413 | 0.265 | 0.2704 | No |
| 30 | Itga2 | 2433 | 0.263 | 0.2779 | No |
| 31 | Cadm2 | 2513 | 0.256 | 0.2802 | No |
| 32 | Amigo1 | 2720 | 0.234 | 0.2715 | No |
| 33 | Ptprc | 2741 | 0.233 | 0.2779 | No |
| 34 | Fbn1 | 2806 | 0.227 | 0.2805 | No |
| 35 | Inppl1 | 2808 | 0.227 | 0.2882 | No |
| 36 | Crat | 2949 | 0.216 | 0.2842 | No |
| 37 | Nlgn3 | 3075 | 0.210 | 0.2812 | No |
| 38 | Icam4 | 3165 | 0.202 | 0.2809 | No |
| 39 | Sorbs3 | 3322 | 0.189 | 0.2747 | No |
| 40 | Arhgef6 | 3440 | 0.178 | 0.2713 | No |
| 41 | Nlgn2 | 3450 | 0.177 | 0.2766 | No |
| 42 | Akt2 | 3473 | 0.175 | 0.2808 | No |
| 43 | Gnai2 | 3675 | 0.159 | 0.2700 | No |
| 44 | Itgb1 | 3793 | 0.150 | 0.2656 | No |
| 45 | Wasl | 3834 | 0.147 | 0.2674 | No |
| 46 | Nectin1 | 3995 | 0.134 | 0.2590 | No |
| 47 | Cdk8 | 4105 | 0.126 | 0.2545 | No |
| 48 | Vwf | 4426 | 0.104 | 0.2321 | No |
| 49 | Vav2 | 4473 | 0.100 | 0.2318 | No |
| 50 | Rhof | 4761 | 0.080 | 0.2112 | No |
| 51 | Rasa1 | 4786 | 0.078 | 0.2119 | No |
| 52 | Pkd1 | 4815 | 0.076 | 0.2123 | No |
| 53 | Nrap | 4980 | 0.065 | 0.2012 | No |
| 54 | Ctnnd1 | 5040 | 0.061 | 0.1985 | No |
| 55 | Bmp1 | 5115 | 0.056 | 0.1944 | No |
| 56 | Cdh1 | 5199 | 0.049 | 0.1894 | No |
| 57 | Cd274 | 5277 | 0.045 | 0.1847 | No |
| 58 | Lima1 | 5879 | 0.007 | 0.1362 | No |
| 59 | Vcan | 6010 | 0.000 | 0.1256 | No |
| 60 | Nectin4 | 6090 | -0.001 | 0.1192 | No |
| 61 | Shc1 | 6141 | -0.003 | 0.1153 | No |
| 62 | Cap1 | 6266 | -0.010 | 0.1056 | No |
| 63 | Syk | 6276 | -0.011 | 0.1052 | No |
| 64 | Dhx16 | 6405 | -0.020 | 0.0955 | No |
| 65 | Myh9 | 6443 | -0.023 | 0.0933 | No |
| 66 | Icam5 | 6476 | -0.025 | 0.0916 | No |
| 67 | Ikbkg | 6592 | -0.030 | 0.0833 | No |
| 68 | Dlg1 | 7188 | -0.067 | 0.0373 | No |
| 69 | Sympk | 7330 | -0.076 | 0.0285 | No |
| 70 | Rac2 | 7355 | -0.077 | 0.0292 | No |
| 71 | Amigo2 | 7473 | -0.084 | 0.0226 | No |
| 72 | Epb41l2 | 7701 | -0.100 | 0.0076 | No |
| 73 | Pik3r3 | 7702 | -0.100 | 0.0110 | No |
| 74 | Pfn1 | 7720 | -0.102 | 0.0131 | No |
| 75 | Mpzl1 | 7819 | -0.110 | 0.0089 | No |
| 76 | Cnn2 | 7933 | -0.117 | 0.0037 | No |
| 77 | Tmem8b | 8208 | -0.135 | -0.0139 | No |
| 78 | Skap2 | 8246 | -0.137 | -0.0122 | No |
| 79 | Itgb4 | 8298 | -0.141 | -0.0115 | No |
| 80 | Akt3 | 8328 | -0.143 | -0.0090 | No |
| 81 | Src | 8355 | -0.145 | -0.0061 | No |
| 82 | Zyx | 8357 | -0.145 | -0.0013 | No |
| 83 | Map4k2 | 8524 | -0.156 | -0.0094 | No |
| 84 | Ldlrap1 | 8670 | -0.169 | -0.0154 | No |
| 85 | Tial1 | 8836 | -0.180 | -0.0226 | No |
| 86 | Nf1 | 8973 | -0.190 | -0.0272 | No |
| 87 | Vasp | 9001 | -0.192 | -0.0228 | No |
| 88 | Cd34 | 9131 | -0.201 | -0.0264 | No |
| 89 | Actg1 | 9316 | -0.216 | -0.0339 | No |
| 90 | Tubg1 | 9360 | -0.218 | -0.0299 | No |
| 91 | Tjp1 | 9383 | -0.220 | -0.0242 | No |
| 92 | Cd86 | 9394 | -0.221 | -0.0175 | No |
| 93 | Sirpa | 9415 | -0.222 | -0.0115 | No |
| 94 | Adam9 | 9451 | -0.225 | -0.0066 | No |
| 95 | Rsu1 | 9528 | -0.227 | -0.0050 | No |
| 96 | Itga10 | 9710 | -0.242 | -0.0114 | No |
| 97 | Ctnna1 | 10157 | -0.279 | -0.0381 | No |
| 98 | Atp1a3 | 10235 | -0.286 | -0.0345 | No |
| 99 | Adam15 | 10236 | -0.286 | -0.0247 | No |
| 100 | Myl12b | 10475 | -0.308 | -0.0335 | No |
| 101 | Actb | 10478 | -0.308 | -0.0231 | No |
| 102 | Plcg1 | 10811 | -0.347 | -0.0382 | No |
| 103 | Tspan4 | 10861 | -0.352 | -0.0301 | No |
| 104 | Actn4 | 10971 | -0.365 | -0.0265 | No |
| 105 | Baiap2 | 11244 | -0.403 | -0.0348 | No |
| 106 | Nf2 | 11273 | -0.407 | -0.0231 | No |
| 107 | Jup | 11482 | -0.437 | -0.0250 | No |
| 108 | Hadh | 11576 | -0.456 | -0.0170 | No |
| 109 | Pecam1 | 11744 | -0.489 | -0.0138 | No |
| 110 | Ywhah | 11925 | -0.543 | -0.0099 | No |
| 111 | Myh10 | 12262 | -0.704 | -0.0130 | No |
| 112 | Icam1 | 12340 | -0.803 | 0.0082 | No |