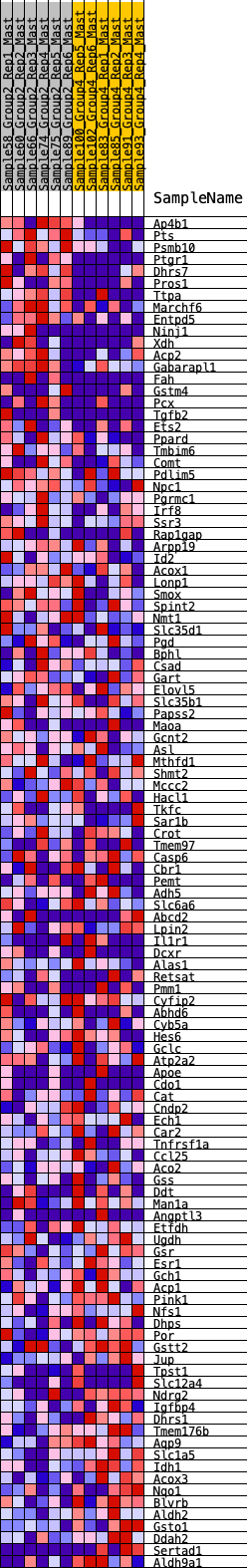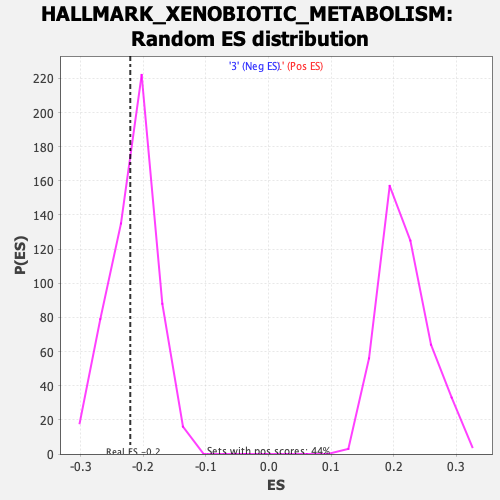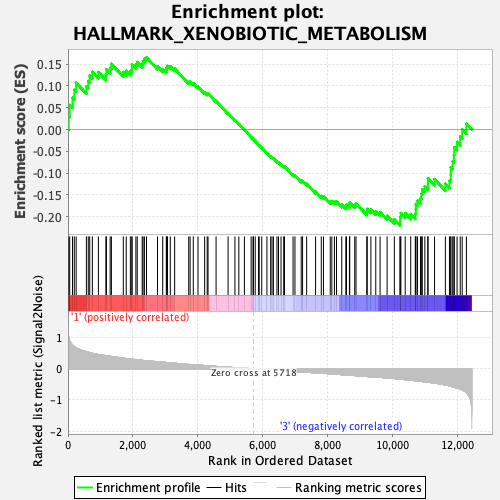
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.2206981 |
| Normalized Enrichment Score (NES) | -1.020725 |
| Nominal p-value | 0.37634408 |
| FDR q-value | 0.69151306 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ap4b1 | 21 | 1.038 | 0.0306 | No |
| 2 | Pts | 52 | 0.905 | 0.0563 | No |
| 3 | Psmb10 | 139 | 0.766 | 0.0731 | No |
| 4 | Ptgr1 | 194 | 0.719 | 0.0910 | No |
| 5 | Dhrs7 | 251 | 0.675 | 0.1075 | No |
| 6 | Pros1 | 566 | 0.552 | 0.0992 | No |
| 7 | Ttpa | 624 | 0.535 | 0.1112 | No |
| 8 | Marchf6 | 669 | 0.517 | 0.1237 | No |
| 9 | Entpd5 | 750 | 0.499 | 0.1327 | No |
| 10 | Ninj1 | 936 | 0.458 | 0.1319 | No |
| 11 | Xdh | 1168 | 0.425 | 0.1264 | No |
| 12 | Acp2 | 1184 | 0.423 | 0.1383 | No |
| 13 | Gabarapl1 | 1299 | 0.403 | 0.1416 | No |
| 14 | Fah | 1337 | 0.397 | 0.1509 | No |
| 15 | Gstm4 | 1704 | 0.345 | 0.1319 | No |
| 16 | Pcx | 1794 | 0.331 | 0.1350 | No |
| 17 | Tgfb2 | 1921 | 0.314 | 0.1345 | No |
| 18 | Ets2 | 1970 | 0.310 | 0.1402 | No |
| 19 | Ppard | 1972 | 0.309 | 0.1498 | No |
| 20 | Tmbim6 | 2092 | 0.295 | 0.1493 | No |
| 21 | Comt | 2133 | 0.290 | 0.1550 | No |
| 22 | Pdlim5 | 2286 | 0.273 | 0.1512 | No |
| 23 | Npc1 | 2312 | 0.270 | 0.1576 | No |
| 24 | Pgrmc1 | 2354 | 0.267 | 0.1625 | No |
| 25 | Irf8 | 2418 | 0.261 | 0.1655 | No |
| 26 | Ssr3 | 2757 | 0.229 | 0.1452 | No |
| 27 | Rap1gap | 2918 | 0.215 | 0.1389 | No |
| 28 | Arpp19 | 3029 | 0.203 | 0.1363 | No |
| 29 | Id2 | 3038 | 0.203 | 0.1420 | No |
| 30 | Acox1 | 3065 | 0.199 | 0.1460 | No |
| 31 | Lonp1 | 3151 | 0.192 | 0.1451 | No |
| 32 | Smox | 3286 | 0.179 | 0.1398 | No |
| 33 | Spint2 | 3719 | 0.142 | 0.1092 | No |
| 34 | Nmt1 | 3762 | 0.138 | 0.1101 | No |
| 35 | Slc35d1 | 3860 | 0.131 | 0.1063 | No |
| 36 | Pgd | 4005 | 0.123 | 0.0984 | No |
| 37 | Bphl | 4210 | 0.106 | 0.0852 | No |
| 38 | Csad | 4288 | 0.099 | 0.0820 | No |
| 39 | Gart | 4314 | 0.097 | 0.0830 | No |
| 40 | Elovl5 | 4562 | 0.078 | 0.0654 | No |
| 41 | Slc35b1 | 4932 | 0.053 | 0.0371 | No |
| 42 | Papss2 | 5143 | 0.040 | 0.0213 | No |
| 43 | Maoa | 5263 | 0.032 | 0.0126 | No |
| 44 | Gcnt2 | 5434 | 0.021 | -0.0005 | No |
| 45 | Asl | 5642 | 0.006 | -0.0171 | No |
| 46 | Mthfd1 | 5700 | 0.002 | -0.0217 | No |
| 47 | Shmt2 | 5713 | 0.000 | -0.0226 | No |
| 48 | Mccc2 | 5768 | -0.001 | -0.0270 | No |
| 49 | Hacl1 | 5871 | -0.010 | -0.0349 | No |
| 50 | Tkfc | 5890 | -0.011 | -0.0361 | No |
| 51 | Sar1b | 5969 | -0.016 | -0.0419 | No |
| 52 | Crot | 6129 | -0.028 | -0.0539 | No |
| 53 | Tmem97 | 6243 | -0.036 | -0.0620 | No |
| 54 | Casp6 | 6285 | -0.039 | -0.0641 | No |
| 55 | Cbr1 | 6328 | -0.042 | -0.0662 | No |
| 56 | Pemt | 6438 | -0.049 | -0.0735 | No |
| 57 | Adh5 | 6485 | -0.052 | -0.0756 | No |
| 58 | Slc6a6 | 6564 | -0.058 | -0.0801 | No |
| 59 | Abcd2 | 6646 | -0.063 | -0.0848 | No |
| 60 | Lpin2 | 6669 | -0.064 | -0.0846 | No |
| 61 | Il1r1 | 6934 | -0.082 | -0.1034 | No |
| 62 | Dcxr | 6992 | -0.086 | -0.1054 | No |
| 63 | Alas1 | 7185 | -0.100 | -0.1178 | No |
| 64 | Retsat | 7227 | -0.103 | -0.1180 | No |
| 65 | Pmm1 | 7353 | -0.111 | -0.1247 | No |
| 66 | Cyfip2 | 7628 | -0.128 | -0.1429 | No |
| 67 | Abhd6 | 7803 | -0.139 | -0.1527 | No |
| 68 | Cyb5a | 7870 | -0.144 | -0.1536 | No |
| 69 | Hes6 | 8079 | -0.159 | -0.1655 | No |
| 70 | Gclc | 8129 | -0.162 | -0.1645 | No |
| 71 | Atp2a2 | 8208 | -0.168 | -0.1655 | No |
| 72 | Apoe | 8273 | -0.172 | -0.1654 | No |
| 73 | Cdo1 | 8434 | -0.185 | -0.1726 | No |
| 74 | Cat | 8560 | -0.192 | -0.1768 | No |
| 75 | Cndp2 | 8580 | -0.194 | -0.1723 | No |
| 76 | Ech1 | 8671 | -0.202 | -0.1733 | No |
| 77 | Car2 | 8680 | -0.202 | -0.1677 | No |
| 78 | Tnfrsf1a | 8834 | -0.214 | -0.1735 | No |
| 79 | Ccl25 | 8873 | -0.217 | -0.1698 | No |
| 80 | Aco2 | 9206 | -0.244 | -0.1891 | No |
| 81 | Gss | 9218 | -0.245 | -0.1824 | No |
| 82 | Ddt | 9332 | -0.253 | -0.1837 | No |
| 83 | Man1a | 9482 | -0.264 | -0.1876 | No |
| 84 | Angptl3 | 9615 | -0.274 | -0.1898 | No |
| 85 | Etfdh | 9831 | -0.291 | -0.1982 | No |
| 86 | Ugdh | 10054 | -0.311 | -0.2065 | No |
| 87 | Gsr | 10230 | -0.329 | -0.2105 | Yes |
| 88 | Esr1 | 10234 | -0.329 | -0.2005 | Yes |
| 89 | Gch1 | 10255 | -0.331 | -0.1918 | Yes |
| 90 | Acp1 | 10389 | -0.347 | -0.1919 | Yes |
| 91 | Pink1 | 10559 | -0.365 | -0.1942 | Yes |
| 92 | Nfs1 | 10699 | -0.380 | -0.1937 | Yes |
| 93 | Dhps | 10715 | -0.383 | -0.1830 | Yes |
| 94 | Por | 10717 | -0.383 | -0.1712 | Yes |
| 95 | Gstt2 | 10769 | -0.388 | -0.1633 | Yes |
| 96 | Jup | 10860 | -0.399 | -0.1582 | Yes |
| 97 | Tpst1 | 10886 | -0.401 | -0.1477 | Yes |
| 98 | Slc12a4 | 10915 | -0.406 | -0.1374 | Yes |
| 99 | Ndrg2 | 10994 | -0.416 | -0.1308 | Yes |
| 100 | Igfbp4 | 11087 | -0.431 | -0.1248 | Yes |
| 101 | Dhrs1 | 11090 | -0.431 | -0.1116 | Yes |
| 102 | Tmem176b | 11293 | -0.458 | -0.1138 | Yes |
| 103 | Aqp9 | 11626 | -0.515 | -0.1247 | Yes |
| 104 | Slc1a5 | 11752 | -0.544 | -0.1179 | Yes |
| 105 | Idh1 | 11793 | -0.557 | -0.1039 | Yes |
| 106 | Acox3 | 11795 | -0.557 | -0.0866 | Yes |
| 107 | Nqo1 | 11853 | -0.575 | -0.0734 | Yes |
| 108 | Blvrb | 11891 | -0.585 | -0.0582 | Yes |
| 109 | Aldh2 | 11901 | -0.589 | -0.0406 | Yes |
| 110 | Gsto1 | 11987 | -0.616 | -0.0284 | Yes |
| 111 | Ddah2 | 12084 | -0.647 | -0.0161 | Yes |
| 112 | Sertad1 | 12145 | -0.677 | 0.0001 | Yes |
| 113 | Aldh9a1 | 12274 | -0.767 | 0.0135 | Yes |