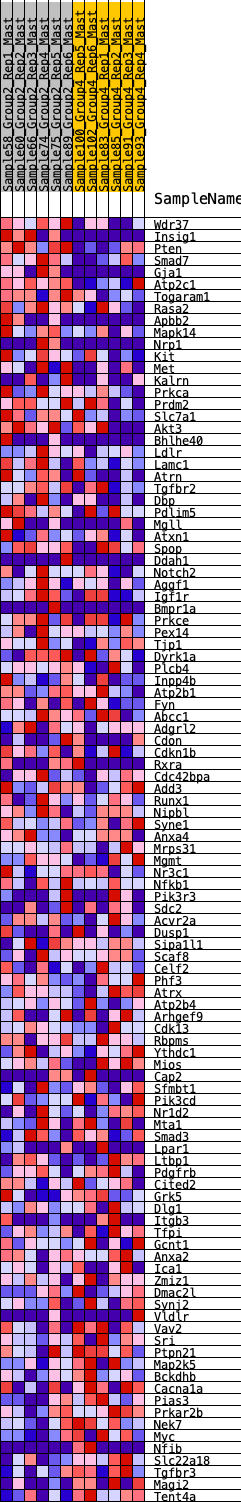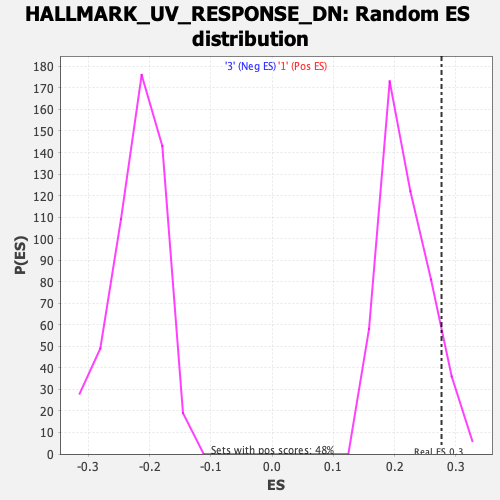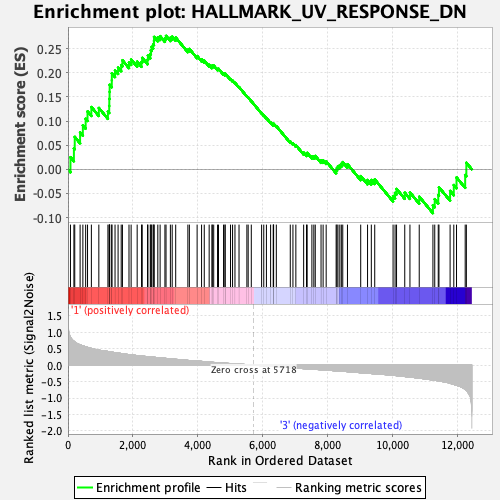
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.2767397 |
| Normalized Enrichment Score (NES) | 1.2695943 |
| Nominal p-value | 0.084033616 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.803 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wdr37 | 76 | 0.851 | 0.0251 | Yes |
| 2 | Insig1 | 182 | 0.727 | 0.0432 | Yes |
| 3 | Pten | 208 | 0.709 | 0.0672 | Yes |
| 4 | Smad7 | 373 | 0.618 | 0.0766 | Yes |
| 5 | Gja1 | 458 | 0.585 | 0.0913 | Yes |
| 6 | Atp2c1 | 544 | 0.556 | 0.1048 | Yes |
| 7 | Togaram1 | 602 | 0.540 | 0.1200 | Yes |
| 8 | Rasa2 | 720 | 0.507 | 0.1292 | Yes |
| 9 | Apbb2 | 950 | 0.453 | 0.1272 | Yes |
| 10 | Mapk14 | 1230 | 0.412 | 0.1198 | Yes |
| 11 | Nrp1 | 1270 | 0.406 | 0.1315 | Yes |
| 12 | Kit | 1273 | 0.406 | 0.1463 | Yes |
| 13 | Met | 1281 | 0.405 | 0.1606 | Yes |
| 14 | Kalrn | 1284 | 0.405 | 0.1753 | Yes |
| 15 | Prkca | 1349 | 0.396 | 0.1846 | Yes |
| 16 | Prdm2 | 1351 | 0.396 | 0.1991 | Yes |
| 17 | Slc7a1 | 1449 | 0.381 | 0.2052 | Yes |
| 18 | Akt3 | 1543 | 0.367 | 0.2111 | Yes |
| 19 | Bhlhe40 | 1639 | 0.352 | 0.2163 | Yes |
| 20 | Ldlr | 1677 | 0.349 | 0.2261 | Yes |
| 21 | Lamc1 | 1879 | 0.319 | 0.2216 | Yes |
| 22 | Atrn | 1945 | 0.313 | 0.2278 | Yes |
| 23 | Tgfbr2 | 2131 | 0.290 | 0.2234 | Yes |
| 24 | Dbp | 2266 | 0.275 | 0.2227 | Yes |
| 25 | Pdlim5 | 2286 | 0.273 | 0.2311 | Yes |
| 26 | Mgll | 2451 | 0.258 | 0.2273 | Yes |
| 27 | Atxn1 | 2462 | 0.256 | 0.2359 | Yes |
| 28 | Spop | 2535 | 0.248 | 0.2392 | Yes |
| 29 | Ddah1 | 2552 | 0.247 | 0.2469 | Yes |
| 30 | Notch2 | 2577 | 0.245 | 0.2540 | Yes |
| 31 | Aggf1 | 2624 | 0.241 | 0.2591 | Yes |
| 32 | Igf1r | 2648 | 0.239 | 0.2660 | Yes |
| 33 | Bmpr1a | 2651 | 0.238 | 0.2746 | Yes |
| 34 | Prkce | 2767 | 0.229 | 0.2737 | Yes |
| 35 | Pex14 | 2838 | 0.223 | 0.2762 | Yes |
| 36 | Tjp1 | 2986 | 0.209 | 0.2719 | Yes |
| 37 | Dyrk1a | 3020 | 0.204 | 0.2767 | Yes |
| 38 | Plcb4 | 3154 | 0.192 | 0.2730 | No |
| 39 | Inpp4b | 3210 | 0.186 | 0.2754 | No |
| 40 | Atp2b1 | 3318 | 0.177 | 0.2732 | No |
| 41 | Fyn | 3693 | 0.144 | 0.2482 | No |
| 42 | Abcc1 | 3739 | 0.140 | 0.2497 | No |
| 43 | Adgrl2 | 3978 | 0.125 | 0.2350 | No |
| 44 | Cdon | 4115 | 0.114 | 0.2281 | No |
| 45 | Cdkn1b | 4198 | 0.107 | 0.2254 | No |
| 46 | Rxra | 4349 | 0.094 | 0.2167 | No |
| 47 | Cdc42bpa | 4436 | 0.088 | 0.2129 | No |
| 48 | Add3 | 4445 | 0.087 | 0.2155 | No |
| 49 | Runx1 | 4489 | 0.083 | 0.2150 | No |
| 50 | Nipbl | 4607 | 0.074 | 0.2083 | No |
| 51 | Syne1 | 4637 | 0.071 | 0.2085 | No |
| 52 | Anxa4 | 4792 | 0.062 | 0.1983 | No |
| 53 | Mrps31 | 4838 | 0.059 | 0.1968 | No |
| 54 | Mgmt | 4839 | 0.059 | 0.1990 | No |
| 55 | Nr3c1 | 5007 | 0.049 | 0.1872 | No |
| 56 | Nfkb1 | 5070 | 0.044 | 0.1838 | No |
| 57 | Pik3r3 | 5144 | 0.040 | 0.1794 | No |
| 58 | Sdc2 | 5269 | 0.032 | 0.1705 | No |
| 59 | Acvr2a | 5511 | 0.016 | 0.1516 | No |
| 60 | Dusp1 | 5551 | 0.013 | 0.1489 | No |
| 61 | Sipa1l1 | 5647 | 0.006 | 0.1414 | No |
| 62 | Scaf8 | 5965 | -0.016 | 0.1163 | No |
| 63 | Celf2 | 6031 | -0.021 | 0.1118 | No |
| 64 | Phf3 | 6113 | -0.027 | 0.1062 | No |
| 65 | Atrx | 6242 | -0.036 | 0.0971 | No |
| 66 | Atp2b4 | 6326 | -0.041 | 0.0919 | No |
| 67 | Arhgef9 | 6327 | -0.041 | 0.0935 | No |
| 68 | Cdk13 | 6335 | -0.042 | 0.0944 | No |
| 69 | Rbpms | 6418 | -0.049 | 0.0896 | No |
| 70 | Ythdc1 | 6848 | -0.077 | 0.0576 | No |
| 71 | Mios | 6930 | -0.082 | 0.0541 | No |
| 72 | Cap2 | 7020 | -0.088 | 0.0501 | No |
| 73 | Sfmbt1 | 7260 | -0.105 | 0.0346 | No |
| 74 | Pik3cd | 7348 | -0.110 | 0.0316 | No |
| 75 | Nr1d2 | 7368 | -0.111 | 0.0341 | No |
| 76 | Mta1 | 7514 | -0.120 | 0.0268 | No |
| 77 | Smad3 | 7574 | -0.124 | 0.0265 | No |
| 78 | Lpar1 | 7618 | -0.128 | 0.0277 | No |
| 79 | Ltbp1 | 7796 | -0.139 | 0.0185 | No |
| 80 | Pdgfrb | 7860 | -0.143 | 0.0186 | No |
| 81 | Cited2 | 7954 | -0.148 | 0.0165 | No |
| 82 | Grk5 | 8261 | -0.172 | -0.0020 | No |
| 83 | Dlg1 | 8284 | -0.174 | 0.0026 | No |
| 84 | Itgb3 | 8317 | -0.176 | 0.0065 | No |
| 85 | Tfpi | 8374 | -0.181 | 0.0086 | No |
| 86 | Gcnt1 | 8421 | -0.184 | 0.0116 | No |
| 87 | Anxa2 | 8465 | -0.186 | 0.0150 | No |
| 88 | Ica1 | 8610 | -0.196 | 0.0105 | No |
| 89 | Zmiz1 | 9018 | -0.228 | -0.0141 | No |
| 90 | Dmac2l | 9229 | -0.246 | -0.0221 | No |
| 91 | Synj2 | 9345 | -0.254 | -0.0221 | No |
| 92 | Vldlr | 9450 | -0.262 | -0.0209 | No |
| 93 | Vav2 | 10015 | -0.307 | -0.0554 | No |
| 94 | Sri | 10077 | -0.314 | -0.0488 | No |
| 95 | Ptpn21 | 10122 | -0.318 | -0.0407 | No |
| 96 | Map2k5 | 10375 | -0.345 | -0.0485 | No |
| 97 | Bckdhb | 10535 | -0.362 | -0.0481 | No |
| 98 | Cacna1a | 10821 | -0.394 | -0.0568 | No |
| 99 | Pias3 | 11243 | -0.449 | -0.0744 | No |
| 100 | Prkar2b | 11299 | -0.459 | -0.0620 | No |
| 101 | Nek7 | 11407 | -0.476 | -0.0532 | No |
| 102 | Myc | 11429 | -0.480 | -0.0373 | No |
| 103 | Nfib | 11772 | -0.551 | -0.0448 | No |
| 104 | Slc22a18 | 11886 | -0.584 | -0.0326 | No |
| 105 | Tgfbr3 | 11970 | -0.610 | -0.0169 | No |
| 106 | Magi2 | 12237 | -0.729 | -0.0117 | No |
| 107 | Tent4a | 12273 | -0.767 | 0.0136 | No |