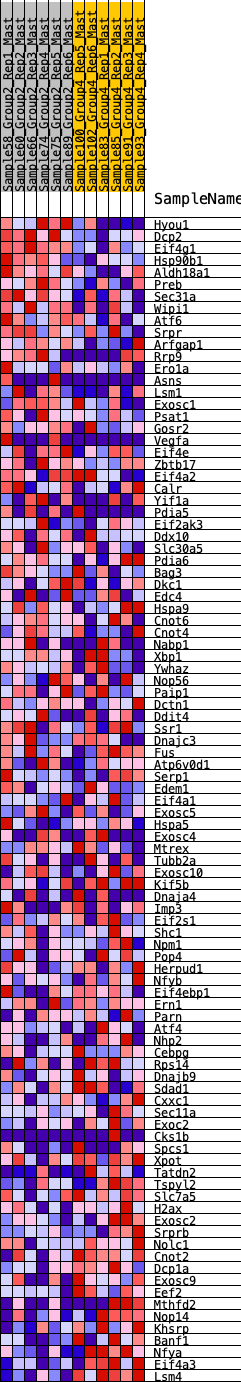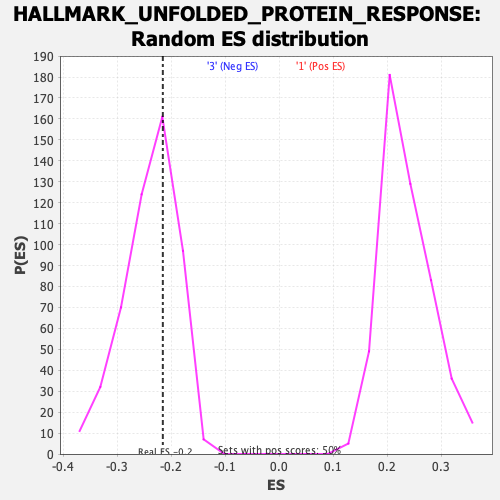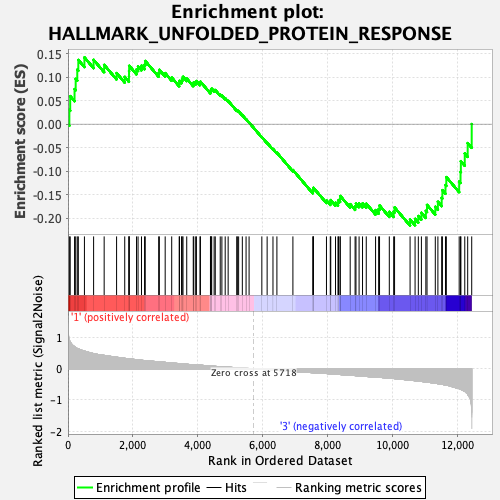
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.21580695 |
| Normalized Enrichment Score (NES) | -0.90330994 |
| Nominal p-value | 0.62151396 |
| FDR q-value | 0.93210465 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hyou1 | 42 | 0.938 | 0.0304 | No |
| 2 | Dcp2 | 68 | 0.866 | 0.0596 | No |
| 3 | Eif4g1 | 202 | 0.714 | 0.0745 | No |
| 4 | Hsp90b1 | 237 | 0.683 | 0.0964 | No |
| 5 | Aldh18a1 | 286 | 0.653 | 0.1160 | No |
| 6 | Preb | 318 | 0.639 | 0.1366 | No |
| 7 | Sec31a | 506 | 0.571 | 0.1420 | No |
| 8 | Wipi1 | 789 | 0.490 | 0.1368 | No |
| 9 | Atf6 | 1114 | 0.432 | 0.1261 | No |
| 10 | Srpr | 1497 | 0.374 | 0.1086 | No |
| 11 | Arfgap1 | 1748 | 0.338 | 0.1005 | No |
| 12 | Rrp9 | 1878 | 0.319 | 0.1016 | No |
| 13 | Ero1a | 1881 | 0.319 | 0.1129 | No |
| 14 | Asns | 1885 | 0.318 | 0.1241 | No |
| 15 | Lsm1 | 2111 | 0.292 | 0.1164 | No |
| 16 | Exosc1 | 2158 | 0.286 | 0.1230 | No |
| 17 | Psat1 | 2264 | 0.276 | 0.1244 | No |
| 18 | Gosr2 | 2359 | 0.266 | 0.1264 | No |
| 19 | Vegfa | 2379 | 0.264 | 0.1344 | No |
| 20 | Eif4e | 2794 | 0.226 | 0.1090 | No |
| 21 | Zbtb17 | 2814 | 0.225 | 0.1156 | No |
| 22 | Eif4a2 | 2994 | 0.208 | 0.1086 | No |
| 23 | Calr | 3195 | 0.187 | 0.0991 | No |
| 24 | Yif1a | 3426 | 0.169 | 0.0866 | No |
| 25 | Pdia5 | 3435 | 0.169 | 0.0920 | No |
| 26 | Eif2ak3 | 3498 | 0.162 | 0.0928 | No |
| 27 | Ddx10 | 3518 | 0.160 | 0.0970 | No |
| 28 | Slc30a5 | 3543 | 0.159 | 0.1008 | No |
| 29 | Pdia6 | 3658 | 0.148 | 0.0969 | No |
| 30 | Bag3 | 3859 | 0.132 | 0.0855 | No |
| 31 | Dkc1 | 3875 | 0.131 | 0.0889 | No |
| 32 | Edc4 | 3935 | 0.128 | 0.0888 | No |
| 33 | Hspa9 | 3956 | 0.126 | 0.0917 | No |
| 34 | Cnot6 | 4067 | 0.118 | 0.0871 | No |
| 35 | Cnot4 | 4080 | 0.117 | 0.0903 | No |
| 36 | Nabp1 | 4389 | 0.091 | 0.0687 | No |
| 37 | Xbp1 | 4402 | 0.090 | 0.0709 | No |
| 38 | Ywhaz | 4408 | 0.090 | 0.0738 | No |
| 39 | Nop56 | 4427 | 0.089 | 0.0755 | No |
| 40 | Paip1 | 4502 | 0.082 | 0.0724 | No |
| 41 | Dctn1 | 4537 | 0.080 | 0.0726 | No |
| 42 | Ddit4 | 4690 | 0.068 | 0.0627 | No |
| 43 | Ssr1 | 4741 | 0.065 | 0.0610 | No |
| 44 | Dnajc3 | 4843 | 0.059 | 0.0549 | No |
| 45 | Fus | 4934 | 0.053 | 0.0495 | No |
| 46 | Atp6v0d1 | 5199 | 0.036 | 0.0295 | No |
| 47 | Serp1 | 5217 | 0.035 | 0.0294 | No |
| 48 | Edem1 | 5257 | 0.033 | 0.0274 | No |
| 49 | Eif4a1 | 5371 | 0.026 | 0.0192 | No |
| 50 | Exosc5 | 5486 | 0.018 | 0.0106 | No |
| 51 | Hspa5 | 5579 | 0.012 | 0.0035 | No |
| 52 | Exosc4 | 5972 | -0.016 | -0.0276 | No |
| 53 | Mtrex | 6135 | -0.028 | -0.0397 | No |
| 54 | Tubb2a | 6315 | -0.041 | -0.0528 | No |
| 55 | Exosc10 | 6436 | -0.049 | -0.0607 | No |
| 56 | Kif5b | 6929 | -0.082 | -0.0976 | No |
| 57 | Dnaja4 | 7543 | -0.122 | -0.1429 | No |
| 58 | Imp3 | 7554 | -0.122 | -0.1393 | No |
| 59 | Eif2s1 | 7562 | -0.123 | -0.1354 | No |
| 60 | Shc1 | 7963 | -0.149 | -0.1625 | No |
| 61 | Npm1 | 8081 | -0.159 | -0.1662 | No |
| 62 | Pop4 | 8095 | -0.160 | -0.1615 | No |
| 63 | Herpud1 | 8242 | -0.170 | -0.1672 | No |
| 64 | Nfyb | 8318 | -0.176 | -0.1669 | No |
| 65 | Eif4ebp1 | 8334 | -0.178 | -0.1617 | No |
| 66 | Ern1 | 8383 | -0.181 | -0.1591 | No |
| 67 | Parn | 8387 | -0.181 | -0.1528 | No |
| 68 | Atf4 | 8694 | -0.203 | -0.1703 | No |
| 69 | Nhp2 | 8845 | -0.214 | -0.1747 | No |
| 70 | Cebpg | 8872 | -0.217 | -0.1690 | No |
| 71 | Rps14 | 8968 | -0.225 | -0.1685 | No |
| 72 | Dnajb9 | 9074 | -0.233 | -0.1687 | No |
| 73 | Sdad1 | 9189 | -0.243 | -0.1692 | No |
| 74 | Cxxc1 | 9476 | -0.263 | -0.1828 | No |
| 75 | Sec11a | 9571 | -0.271 | -0.1807 | No |
| 76 | Exoc2 | 9598 | -0.273 | -0.1730 | No |
| 77 | Cks1b | 9899 | -0.297 | -0.1866 | No |
| 78 | Spcs1 | 10034 | -0.309 | -0.1863 | No |
| 79 | Xpot | 10061 | -0.312 | -0.1772 | No |
| 80 | Tatdn2 | 10539 | -0.362 | -0.2028 | Yes |
| 81 | Tspyl2 | 10693 | -0.379 | -0.2015 | Yes |
| 82 | Slc7a5 | 10793 | -0.391 | -0.1954 | Yes |
| 83 | H2ax | 10888 | -0.402 | -0.1885 | Yes |
| 84 | Exosc2 | 11025 | -0.422 | -0.1844 | Yes |
| 85 | Srprb | 11061 | -0.427 | -0.1718 | Yes |
| 86 | Nolc1 | 11316 | -0.462 | -0.1758 | Yes |
| 87 | Cnot2 | 11396 | -0.475 | -0.1651 | Yes |
| 88 | Dcp1a | 11513 | -0.493 | -0.1567 | Yes |
| 89 | Exosc9 | 11535 | -0.497 | -0.1405 | Yes |
| 90 | Eef2 | 11630 | -0.516 | -0.1295 | Yes |
| 91 | Mthfd2 | 11656 | -0.522 | -0.1127 | Yes |
| 92 | Nop14 | 12052 | -0.634 | -0.1219 | Yes |
| 93 | Khsrp | 12094 | -0.650 | -0.1017 | Yes |
| 94 | Banf1 | 12104 | -0.658 | -0.0788 | Yes |
| 95 | Nfya | 12224 | -0.721 | -0.0624 | Yes |
| 96 | Eif4a3 | 12314 | -0.814 | -0.0403 | Yes |
| 97 | Lsm4 | 12438 | -1.402 | 0.0002 | Yes |