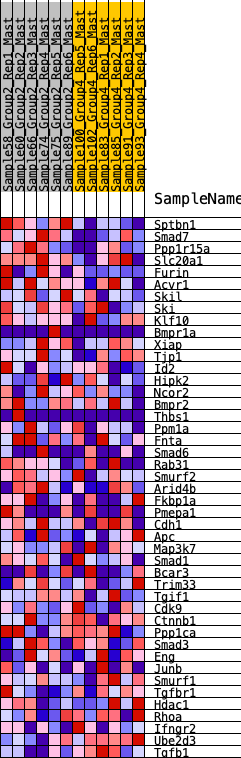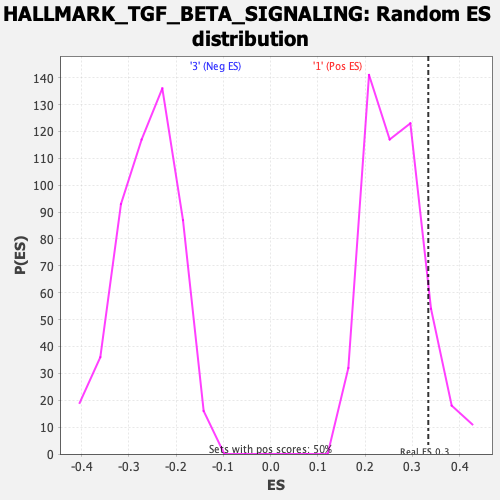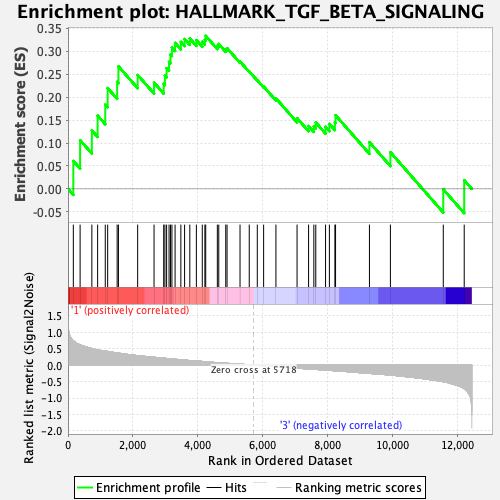
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.3335646 |
| Normalized Enrichment Score (NES) | 1.2643049 |
| Nominal p-value | 0.14112903 |
| FDR q-value | 0.93497586 |
| FWER p-Value | 0.808 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sptbn1 | 164 | 0.737 | 0.0609 | Yes |
| 2 | Smad7 | 373 | 0.618 | 0.1062 | Yes |
| 3 | Ppp1r15a | 735 | 0.502 | 0.1275 | Yes |
| 4 | Slc20a1 | 913 | 0.463 | 0.1597 | Yes |
| 5 | Furin | 1148 | 0.427 | 0.1838 | Yes |
| 6 | Acvr1 | 1222 | 0.415 | 0.2197 | Yes |
| 7 | Skil | 1514 | 0.372 | 0.2336 | Yes |
| 8 | Ski | 1554 | 0.365 | 0.2671 | Yes |
| 9 | Klf10 | 2146 | 0.288 | 0.2483 | Yes |
| 10 | Bmpr1a | 2651 | 0.238 | 0.2316 | Yes |
| 11 | Xiap | 2948 | 0.212 | 0.2291 | Yes |
| 12 | Tjp1 | 2986 | 0.209 | 0.2471 | Yes |
| 13 | Id2 | 3038 | 0.203 | 0.2633 | Yes |
| 14 | Hipk2 | 3113 | 0.197 | 0.2771 | Yes |
| 15 | Ncor2 | 3157 | 0.191 | 0.2929 | Yes |
| 16 | Bmpr2 | 3202 | 0.187 | 0.3081 | Yes |
| 17 | Thbs1 | 3299 | 0.178 | 0.3183 | Yes |
| 18 | Ppm1a | 3477 | 0.164 | 0.3205 | Yes |
| 19 | Fnta | 3592 | 0.154 | 0.3268 | Yes |
| 20 | Smad6 | 3753 | 0.139 | 0.3278 | Yes |
| 21 | Rab31 | 3955 | 0.127 | 0.3243 | Yes |
| 22 | Smurf2 | 4137 | 0.112 | 0.3209 | Yes |
| 23 | Arid4b | 4217 | 0.106 | 0.3252 | Yes |
| 24 | Fkbp1a | 4243 | 0.104 | 0.3336 | Yes |
| 25 | Pmepa1 | 4601 | 0.074 | 0.3122 | No |
| 26 | Cdh1 | 4645 | 0.071 | 0.3159 | No |
| 27 | Apc | 4860 | 0.058 | 0.3044 | No |
| 28 | Map3k7 | 4900 | 0.055 | 0.3069 | No |
| 29 | Smad1 | 5300 | 0.030 | 0.2777 | No |
| 30 | Bcar3 | 5584 | 0.011 | 0.2560 | No |
| 31 | Trim33 | 5832 | -0.007 | 0.2368 | No |
| 32 | Tgif1 | 6027 | -0.020 | 0.2231 | No |
| 33 | Cdk9 | 6406 | -0.048 | 0.1974 | No |
| 34 | Ctnnb1 | 7057 | -0.091 | 0.1541 | No |
| 35 | Ppp1ca | 7412 | -0.114 | 0.1371 | No |
| 36 | Smad3 | 7574 | -0.124 | 0.1365 | No |
| 37 | Eng | 7634 | -0.129 | 0.1447 | No |
| 38 | Junb | 7935 | -0.147 | 0.1353 | No |
| 39 | Smurf1 | 8053 | -0.157 | 0.1416 | No |
| 40 | Tgfbr1 | 8223 | -0.169 | 0.1450 | No |
| 41 | Hdac1 | 8243 | -0.170 | 0.1606 | No |
| 42 | Rhoa | 9287 | -0.251 | 0.1017 | No |
| 43 | Ifngr2 | 9933 | -0.301 | 0.0799 | No |
| 44 | Ube2d3 | 11562 | -0.503 | -0.0009 | No |
| 45 | Tgfb1 | 12207 | -0.714 | 0.0189 | No |