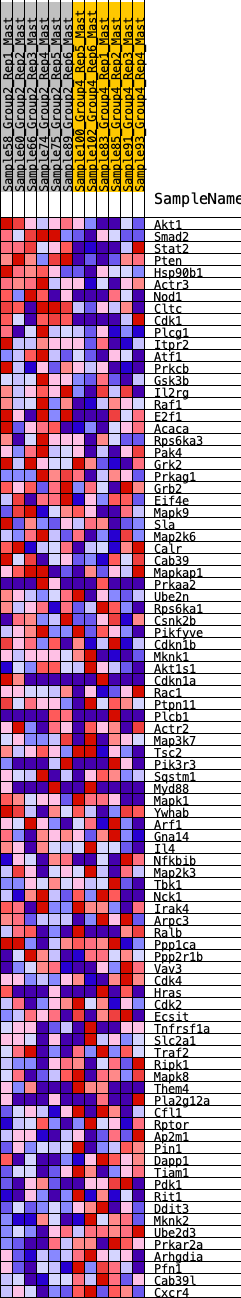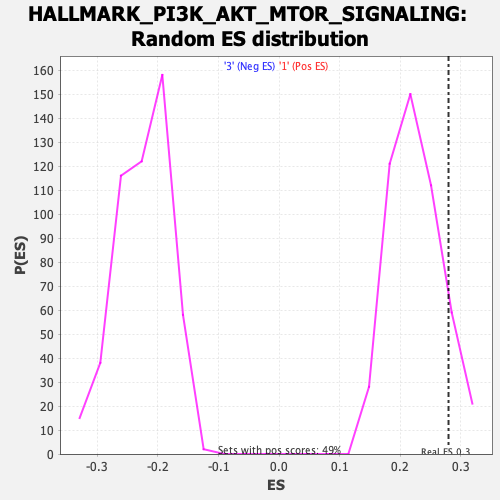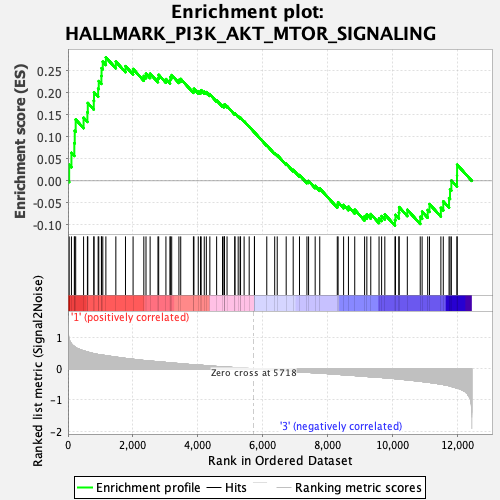
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.27970383 |
| Normalized Enrichment Score (NES) | 1.2504991 |
| Nominal p-value | 0.087576374 |
| FDR q-value | 0.7600701 |
| FWER p-Value | 0.818 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 36 | 0.968 | 0.0364 | Yes |
| 2 | Smad2 | 106 | 0.804 | 0.0635 | Yes |
| 3 | Stat2 | 195 | 0.718 | 0.0856 | Yes |
| 4 | Pten | 208 | 0.709 | 0.1134 | Yes |
| 5 | Hsp90b1 | 237 | 0.683 | 0.1390 | Yes |
| 6 | Actr3 | 480 | 0.580 | 0.1429 | Yes |
| 7 | Nod1 | 598 | 0.541 | 0.1555 | Yes |
| 8 | Cltc | 610 | 0.539 | 0.1765 | Yes |
| 9 | Cdk1 | 794 | 0.488 | 0.1815 | Yes |
| 10 | Plcg1 | 802 | 0.486 | 0.2007 | Yes |
| 11 | Itpr2 | 929 | 0.460 | 0.2091 | Yes |
| 12 | Atf1 | 948 | 0.454 | 0.2261 | Yes |
| 13 | Prkcb | 1029 | 0.443 | 0.2377 | Yes |
| 14 | Gsk3b | 1034 | 0.442 | 0.2553 | Yes |
| 15 | Il2rg | 1072 | 0.436 | 0.2700 | Yes |
| 16 | Raf1 | 1167 | 0.425 | 0.2797 | Yes |
| 17 | E2f1 | 1474 | 0.378 | 0.2703 | No |
| 18 | Acaca | 1771 | 0.334 | 0.2599 | No |
| 19 | Rps6ka3 | 2005 | 0.307 | 0.2535 | No |
| 20 | Pak4 | 2336 | 0.269 | 0.2377 | No |
| 21 | Grk2 | 2403 | 0.263 | 0.2430 | No |
| 22 | Prkag1 | 2530 | 0.249 | 0.2430 | No |
| 23 | Grb2 | 2772 | 0.228 | 0.2327 | No |
| 24 | Eif4e | 2794 | 0.226 | 0.2402 | No |
| 25 | Mapk9 | 3017 | 0.205 | 0.2306 | No |
| 26 | Sla | 3143 | 0.193 | 0.2283 | No |
| 27 | Map2k6 | 3152 | 0.192 | 0.2355 | No |
| 28 | Calr | 3195 | 0.187 | 0.2397 | No |
| 29 | Cab39 | 3417 | 0.170 | 0.2287 | No |
| 30 | Mapkap1 | 3473 | 0.164 | 0.2309 | No |
| 31 | Prkaa2 | 3864 | 0.131 | 0.2047 | No |
| 32 | Ube2n | 3880 | 0.130 | 0.2087 | No |
| 33 | Rps6ka1 | 4013 | 0.122 | 0.2030 | No |
| 34 | Csnk2b | 4082 | 0.117 | 0.2023 | No |
| 35 | Pikfyve | 4102 | 0.115 | 0.2054 | No |
| 36 | Cdkn1b | 4198 | 0.107 | 0.2021 | No |
| 37 | Mknk1 | 4262 | 0.101 | 0.2011 | No |
| 38 | Akt1s1 | 4370 | 0.093 | 0.1962 | No |
| 39 | Cdkn1a | 4579 | 0.077 | 0.1825 | No |
| 40 | Rac1 | 4760 | 0.064 | 0.1705 | No |
| 41 | Ptpn11 | 4809 | 0.061 | 0.1691 | No |
| 42 | Plcb1 | 4812 | 0.061 | 0.1714 | No |
| 43 | Actr2 | 4821 | 0.060 | 0.1732 | No |
| 44 | Map3k7 | 4900 | 0.055 | 0.1691 | No |
| 45 | Tsc2 | 5138 | 0.040 | 0.1516 | No |
| 46 | Pik3r3 | 5144 | 0.040 | 0.1528 | No |
| 47 | Sqstm1 | 5243 | 0.034 | 0.1462 | No |
| 48 | Myd88 | 5302 | 0.030 | 0.1428 | No |
| 49 | Mapk1 | 5316 | 0.029 | 0.1429 | No |
| 50 | Ywhab | 5426 | 0.022 | 0.1350 | No |
| 51 | Arf1 | 5582 | 0.011 | 0.1229 | No |
| 52 | Gna14 | 5743 | -0.000 | 0.1100 | No |
| 53 | Il4 | 5746 | -0.000 | 0.1098 | No |
| 54 | Nfkbib | 6124 | -0.028 | 0.0804 | No |
| 55 | Map2k3 | 6368 | -0.045 | 0.0626 | No |
| 56 | Tbk1 | 6446 | -0.050 | 0.0584 | No |
| 57 | Nck1 | 6722 | -0.069 | 0.0389 | No |
| 58 | Irak4 | 6938 | -0.082 | 0.0249 | No |
| 59 | Arpc3 | 7132 | -0.097 | 0.0132 | No |
| 60 | Ralb | 7361 | -0.111 | -0.0008 | No |
| 61 | Ppp1ca | 7412 | -0.114 | -0.0002 | No |
| 62 | Ppp2r1b | 7615 | -0.128 | -0.0113 | No |
| 63 | Vav3 | 7756 | -0.136 | -0.0172 | No |
| 64 | Cdk4 | 8296 | -0.174 | -0.0537 | No |
| 65 | Hras | 8321 | -0.177 | -0.0485 | No |
| 66 | Cdk2 | 8491 | -0.188 | -0.0545 | No |
| 67 | Ecsit | 8640 | -0.199 | -0.0584 | No |
| 68 | Tnfrsf1a | 8834 | -0.214 | -0.0653 | No |
| 69 | Slc2a1 | 9138 | -0.239 | -0.0801 | No |
| 70 | Traf2 | 9207 | -0.244 | -0.0757 | No |
| 71 | Ripk1 | 9328 | -0.253 | -0.0752 | No |
| 72 | Mapk8 | 9583 | -0.272 | -0.0847 | No |
| 73 | Them4 | 9662 | -0.277 | -0.0797 | No |
| 74 | Pla2g12a | 9761 | -0.285 | -0.0761 | No |
| 75 | Cfl1 | 10074 | -0.313 | -0.0886 | No |
| 76 | Rptor | 10090 | -0.314 | -0.0770 | No |
| 77 | Ap2m1 | 10194 | -0.325 | -0.0722 | No |
| 78 | Pin1 | 10202 | -0.326 | -0.0595 | No |
| 79 | Dapp1 | 10454 | -0.352 | -0.0655 | No |
| 80 | Tiam1 | 10852 | -0.398 | -0.0815 | No |
| 81 | Pdk1 | 10910 | -0.406 | -0.0696 | No |
| 82 | Rit1 | 11082 | -0.430 | -0.0660 | No |
| 83 | Ddit3 | 11137 | -0.437 | -0.0526 | No |
| 84 | Mknk2 | 11488 | -0.489 | -0.0610 | No |
| 85 | Ube2d3 | 11562 | -0.503 | -0.0465 | No |
| 86 | Prkar2a | 11740 | -0.540 | -0.0389 | No |
| 87 | Arhgdia | 11770 | -0.550 | -0.0189 | No |
| 88 | Pfn1 | 11811 | -0.563 | 0.0007 | No |
| 89 | Cab39l | 11985 | -0.615 | 0.0117 | No |
| 90 | Cxcr4 | 11988 | -0.617 | 0.0367 | No |