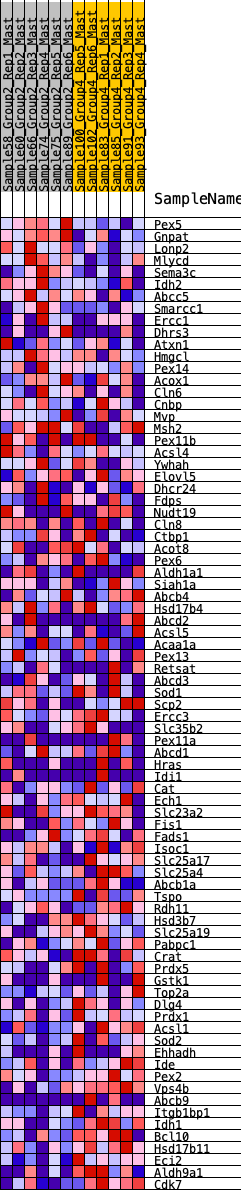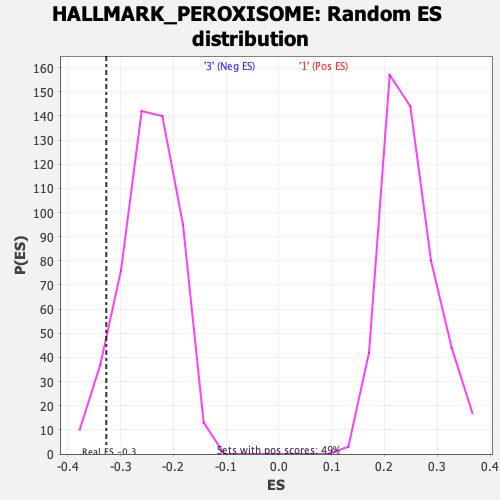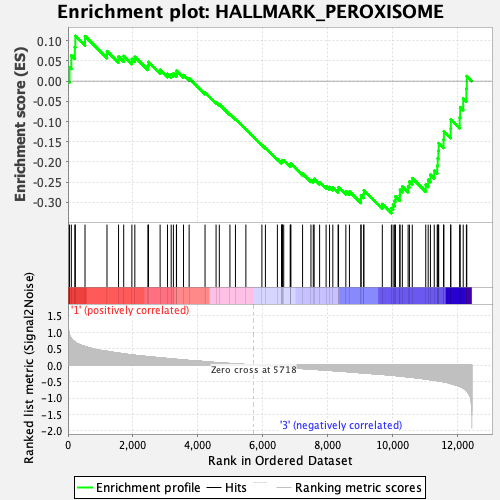
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.3269888 |
| Normalized Enrichment Score (NES) | -1.3361032 |
| Nominal p-value | 0.0662768 |
| FDR q-value | 0.54711103 |
| FWER p-Value | 0.699 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 47 | 0.919 | 0.0345 | No |
| 2 | Gnpat | 103 | 0.807 | 0.0637 | No |
| 3 | Lonp2 | 211 | 0.706 | 0.0844 | No |
| 4 | Mlycd | 226 | 0.690 | 0.1120 | No |
| 5 | Sema3c | 524 | 0.564 | 0.1115 | No |
| 6 | Idh2 | 1201 | 0.420 | 0.0743 | No |
| 7 | Abcc5 | 1557 | 0.365 | 0.0608 | No |
| 8 | Smarcc1 | 1716 | 0.343 | 0.0623 | No |
| 9 | Ercc1 | 1968 | 0.310 | 0.0549 | No |
| 10 | Dhrs3 | 2059 | 0.300 | 0.0601 | No |
| 11 | Atxn1 | 2462 | 0.256 | 0.0383 | No |
| 12 | Hmgcl | 2480 | 0.255 | 0.0475 | No |
| 13 | Pex14 | 2838 | 0.223 | 0.0279 | No |
| 14 | Acox1 | 3065 | 0.199 | 0.0179 | No |
| 15 | Cln6 | 3179 | 0.189 | 0.0167 | No |
| 16 | Cnbp | 3247 | 0.183 | 0.0189 | No |
| 17 | Mvp | 3340 | 0.175 | 0.0188 | No |
| 18 | Msh2 | 3344 | 0.175 | 0.0258 | No |
| 19 | Pex11b | 3559 | 0.158 | 0.0151 | No |
| 20 | Acsl4 | 3732 | 0.140 | 0.0070 | No |
| 21 | Ywhah | 4222 | 0.105 | -0.0282 | No |
| 22 | Elovl5 | 4562 | 0.078 | -0.0524 | No |
| 23 | Dhcr24 | 4662 | 0.070 | -0.0575 | No |
| 24 | Fdps | 4989 | 0.050 | -0.0818 | No |
| 25 | Nudt19 | 5159 | 0.039 | -0.0938 | No |
| 26 | Cln8 | 5480 | 0.018 | -0.1190 | No |
| 27 | Ctbp1 | 5974 | -0.016 | -0.1582 | No |
| 28 | Acot8 | 6082 | -0.025 | -0.1658 | No |
| 29 | Pex6 | 6451 | -0.050 | -0.1935 | No |
| 30 | Aldh1a1 | 6578 | -0.058 | -0.2012 | No |
| 31 | Siah1a | 6588 | -0.059 | -0.1995 | No |
| 32 | Abcb4 | 6604 | -0.060 | -0.1982 | No |
| 33 | Hsd17b4 | 6606 | -0.060 | -0.1958 | No |
| 34 | Abcd2 | 6646 | -0.063 | -0.1963 | No |
| 35 | Acsl5 | 6846 | -0.077 | -0.2092 | No |
| 36 | Acaa1a | 6859 | -0.078 | -0.2069 | No |
| 37 | Pex13 | 6865 | -0.078 | -0.2041 | No |
| 38 | Retsat | 7227 | -0.103 | -0.2290 | No |
| 39 | Abcd3 | 7485 | -0.118 | -0.2449 | No |
| 40 | Sod1 | 7557 | -0.122 | -0.2455 | No |
| 41 | Scp2 | 7583 | -0.125 | -0.2423 | No |
| 42 | Ercc3 | 7752 | -0.135 | -0.2503 | No |
| 43 | Slc35b2 | 7952 | -0.148 | -0.2602 | No |
| 44 | Pex11a | 8058 | -0.157 | -0.2622 | No |
| 45 | Abcd1 | 8159 | -0.164 | -0.2634 | No |
| 46 | Hras | 8321 | -0.177 | -0.2691 | No |
| 47 | Idi1 | 8335 | -0.178 | -0.2627 | No |
| 48 | Cat | 8560 | -0.192 | -0.2728 | No |
| 49 | Ech1 | 8671 | -0.202 | -0.2733 | No |
| 50 | Slc23a2 | 9025 | -0.229 | -0.2924 | No |
| 51 | Fis1 | 9027 | -0.229 | -0.2829 | No |
| 52 | Fads1 | 9109 | -0.236 | -0.2796 | No |
| 53 | Isoc1 | 9116 | -0.237 | -0.2703 | No |
| 54 | Slc25a17 | 9684 | -0.279 | -0.3045 | No |
| 55 | Slc25a4 | 9963 | -0.303 | -0.3144 | Yes |
| 56 | Abcb1a | 10016 | -0.307 | -0.3058 | Yes |
| 57 | Tspo | 10063 | -0.312 | -0.2965 | Yes |
| 58 | Rdh11 | 10089 | -0.314 | -0.2854 | Yes |
| 59 | Hsd3b7 | 10222 | -0.328 | -0.2824 | Yes |
| 60 | Slc25a19 | 10228 | -0.328 | -0.2692 | Yes |
| 61 | Pabpc1 | 10301 | -0.336 | -0.2610 | Yes |
| 62 | Crat | 10479 | -0.354 | -0.2605 | Yes |
| 63 | Prdx5 | 10519 | -0.360 | -0.2487 | Yes |
| 64 | Gstk1 | 10607 | -0.369 | -0.2404 | Yes |
| 65 | Top2a | 11022 | -0.421 | -0.2563 | Yes |
| 66 | Dlg4 | 11097 | -0.431 | -0.2443 | Yes |
| 67 | Prdx1 | 11166 | -0.440 | -0.2315 | Yes |
| 68 | Acsl1 | 11284 | -0.457 | -0.2220 | Yes |
| 69 | Sod2 | 11375 | -0.471 | -0.2096 | Yes |
| 70 | Ehhadh | 11391 | -0.474 | -0.1911 | Yes |
| 71 | Ide | 11411 | -0.477 | -0.1728 | Yes |
| 72 | Pex2 | 11421 | -0.479 | -0.1536 | Yes |
| 73 | Vps4b | 11570 | -0.505 | -0.1445 | Yes |
| 74 | Abcb9 | 11581 | -0.506 | -0.1243 | Yes |
| 75 | Itgb1bp1 | 11790 | -0.556 | -0.1179 | Yes |
| 76 | Idh1 | 11793 | -0.557 | -0.0949 | Yes |
| 77 | Bcl10 | 12067 | -0.641 | -0.0903 | Yes |
| 78 | Hsd17b11 | 12083 | -0.647 | -0.0645 | Yes |
| 79 | Eci2 | 12176 | -0.696 | -0.0430 | Yes |
| 80 | Aldh9a1 | 12274 | -0.767 | -0.0189 | Yes |
| 81 | Cdk7 | 12286 | -0.776 | 0.0125 | Yes |