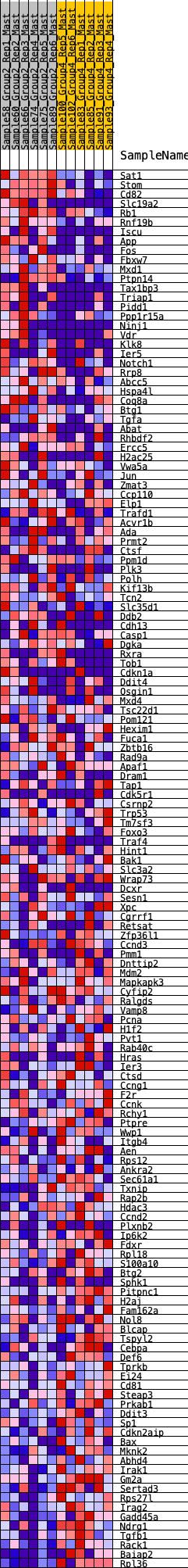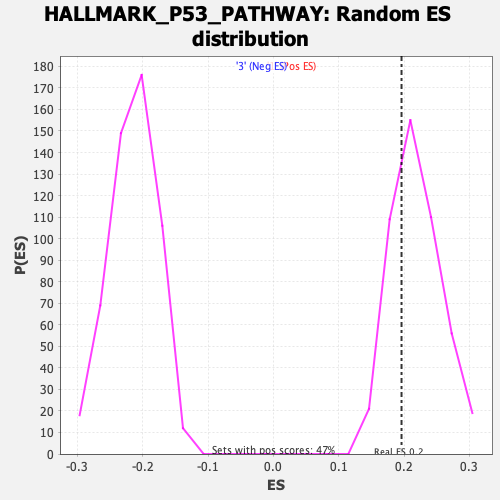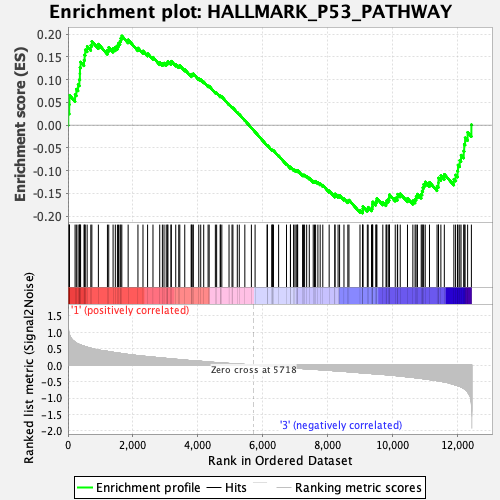
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_P53_PATHWAY |
| Enrichment Score (ES) | 0.19617808 |
| Normalized Enrichment Score (NES) | 0.90371597 |
| Nominal p-value | 0.6957447 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sat1 | 8 | 1.214 | 0.0265 | Yes |
| 2 | Stom | 38 | 0.953 | 0.0455 | Yes |
| 3 | Cd82 | 45 | 0.922 | 0.0657 | Yes |
| 4 | Slc19a2 | 218 | 0.697 | 0.0673 | Yes |
| 5 | Rb1 | 257 | 0.670 | 0.0792 | Yes |
| 6 | Rnf19b | 310 | 0.641 | 0.0893 | Yes |
| 7 | Iscu | 351 | 0.627 | 0.1001 | Yes |
| 8 | App | 366 | 0.620 | 0.1129 | Yes |
| 9 | Fos | 369 | 0.619 | 0.1266 | Yes |
| 10 | Fbxw7 | 388 | 0.611 | 0.1388 | Yes |
| 11 | Mxd1 | 494 | 0.574 | 0.1431 | Yes |
| 12 | Ptpn14 | 512 | 0.569 | 0.1545 | Yes |
| 13 | Tax1bp3 | 531 | 0.562 | 0.1656 | Yes |
| 14 | Triap1 | 590 | 0.544 | 0.1731 | Yes |
| 15 | Pidd1 | 703 | 0.513 | 0.1754 | Yes |
| 16 | Ppp1r15a | 735 | 0.502 | 0.1842 | Yes |
| 17 | Ninj1 | 936 | 0.458 | 0.1781 | Yes |
| 18 | Vdr | 1215 | 0.418 | 0.1649 | Yes |
| 19 | Klk8 | 1255 | 0.409 | 0.1709 | Yes |
| 20 | Ier5 | 1389 | 0.389 | 0.1688 | Yes |
| 21 | Notch1 | 1458 | 0.379 | 0.1717 | Yes |
| 22 | Rrp8 | 1523 | 0.371 | 0.1748 | Yes |
| 23 | Abcc5 | 1557 | 0.365 | 0.1803 | Yes |
| 24 | Hspa4l | 1603 | 0.359 | 0.1847 | Yes |
| 25 | Coq8a | 1626 | 0.355 | 0.1909 | Yes |
| 26 | Btg1 | 1658 | 0.350 | 0.1962 | Yes |
| 27 | Tgfa | 1853 | 0.323 | 0.1876 | No |
| 28 | Abat | 2155 | 0.287 | 0.1696 | No |
| 29 | Rhbdf2 | 2310 | 0.271 | 0.1631 | No |
| 30 | Ercc5 | 2452 | 0.258 | 0.1574 | No |
| 31 | H2ac25 | 2618 | 0.241 | 0.1494 | No |
| 32 | Vwa5a | 2826 | 0.224 | 0.1376 | No |
| 33 | Jun | 2908 | 0.216 | 0.1358 | No |
| 34 | Zmat3 | 2958 | 0.211 | 0.1365 | No |
| 35 | Ccp110 | 3026 | 0.203 | 0.1356 | No |
| 36 | Elp1 | 3060 | 0.200 | 0.1374 | No |
| 37 | Trafd1 | 3079 | 0.198 | 0.1404 | No |
| 38 | Acvr1b | 3171 | 0.190 | 0.1373 | No |
| 39 | Ada | 3183 | 0.189 | 0.1406 | No |
| 40 | Prmt2 | 3319 | 0.177 | 0.1336 | No |
| 41 | Ctsf | 3412 | 0.171 | 0.1299 | No |
| 42 | Ppm1d | 3444 | 0.168 | 0.1311 | No |
| 43 | Plk3 | 3597 | 0.154 | 0.1222 | No |
| 44 | Polh | 3798 | 0.136 | 0.1090 | No |
| 45 | Kif13b | 3800 | 0.136 | 0.1120 | No |
| 46 | Tcn2 | 3845 | 0.133 | 0.1114 | No |
| 47 | Slc35d1 | 3860 | 0.131 | 0.1132 | No |
| 48 | Ddb2 | 4029 | 0.120 | 0.1022 | No |
| 49 | Cdh13 | 4089 | 0.116 | 0.1000 | No |
| 50 | Casp1 | 4181 | 0.109 | 0.0950 | No |
| 51 | Dgka | 4316 | 0.096 | 0.0863 | No |
| 52 | Rxra | 4349 | 0.094 | 0.0858 | No |
| 53 | Tob1 | 4540 | 0.080 | 0.0721 | No |
| 54 | Cdkn1a | 4579 | 0.077 | 0.0707 | No |
| 55 | Ddit4 | 4690 | 0.068 | 0.0633 | No |
| 56 | Osgin1 | 4695 | 0.068 | 0.0645 | No |
| 57 | Mxd4 | 4739 | 0.065 | 0.0625 | No |
| 58 | Tsc22d1 | 4961 | 0.051 | 0.0457 | No |
| 59 | Pom121 | 5053 | 0.046 | 0.0393 | No |
| 60 | Hexim1 | 5081 | 0.044 | 0.0381 | No |
| 61 | Fuca1 | 5216 | 0.035 | 0.0279 | No |
| 62 | Zbtb16 | 5277 | 0.031 | 0.0238 | No |
| 63 | Rad9a | 5448 | 0.020 | 0.0104 | No |
| 64 | Apaf1 | 5650 | 0.006 | -0.0058 | No |
| 65 | Dram1 | 5764 | -0.001 | -0.0150 | No |
| 66 | Tap1 | 6134 | -0.028 | -0.0444 | No |
| 67 | Cdk5r1 | 6144 | -0.029 | -0.0444 | No |
| 68 | Csrnp2 | 6277 | -0.038 | -0.0543 | No |
| 69 | Trp53 | 6295 | -0.039 | -0.0548 | No |
| 70 | Tm7sf3 | 6306 | -0.040 | -0.0547 | No |
| 71 | Foxo3 | 6330 | -0.042 | -0.0557 | No |
| 72 | Traf4 | 6489 | -0.052 | -0.0674 | No |
| 73 | Hint1 | 6732 | -0.069 | -0.0855 | No |
| 74 | Bak1 | 6853 | -0.077 | -0.0935 | No |
| 75 | Slc3a2 | 6854 | -0.078 | -0.0918 | No |
| 76 | Wrap73 | 6950 | -0.083 | -0.0976 | No |
| 77 | Dcxr | 6992 | -0.086 | -0.0990 | No |
| 78 | Sesn1 | 7032 | -0.089 | -0.1002 | No |
| 79 | Xpc | 7066 | -0.091 | -0.1009 | No |
| 80 | Cgrrf1 | 7076 | -0.092 | -0.0995 | No |
| 81 | Retsat | 7227 | -0.103 | -0.1094 | No |
| 82 | Zfp36l1 | 7257 | -0.105 | -0.1095 | No |
| 83 | Ccnd3 | 7289 | -0.106 | -0.1096 | No |
| 84 | Pmm1 | 7353 | -0.111 | -0.1122 | No |
| 85 | Dnttip2 | 7434 | -0.116 | -0.1162 | No |
| 86 | Mdm2 | 7555 | -0.122 | -0.1232 | No |
| 87 | Mapkapk3 | 7594 | -0.126 | -0.1235 | No |
| 88 | Cyfip2 | 7628 | -0.128 | -0.1233 | No |
| 89 | Ralgds | 7699 | -0.133 | -0.1260 | No |
| 90 | Vamp8 | 7769 | -0.137 | -0.1285 | No |
| 91 | Pcna | 7850 | -0.142 | -0.1319 | No |
| 92 | H1f2 | 8046 | -0.156 | -0.1442 | No |
| 93 | Pvt1 | 8222 | -0.169 | -0.1547 | No |
| 94 | Rab40c | 8225 | -0.169 | -0.1511 | No |
| 95 | Hras | 8321 | -0.177 | -0.1548 | No |
| 96 | Ier3 | 8367 | -0.180 | -0.1545 | No |
| 97 | Ctsd | 8502 | -0.189 | -0.1611 | No |
| 98 | Ccng1 | 8622 | -0.197 | -0.1664 | No |
| 99 | F2r | 8656 | -0.200 | -0.1646 | No |
| 100 | Ccnk | 8996 | -0.227 | -0.1871 | No |
| 101 | Rchy1 | 9075 | -0.233 | -0.1882 | No |
| 102 | Ptpre | 9082 | -0.233 | -0.1835 | No |
| 103 | Wwp1 | 9086 | -0.234 | -0.1785 | No |
| 104 | Itgb4 | 9222 | -0.246 | -0.1840 | No |
| 105 | Aen | 9247 | -0.248 | -0.1804 | No |
| 106 | Rps12 | 9351 | -0.255 | -0.1830 | No |
| 107 | Ankra2 | 9373 | -0.256 | -0.1790 | No |
| 108 | Sec61a1 | 9380 | -0.257 | -0.1738 | No |
| 109 | Txnip | 9389 | -0.257 | -0.1686 | No |
| 110 | Rap2b | 9483 | -0.264 | -0.1703 | No |
| 111 | Hdac3 | 9500 | -0.266 | -0.1656 | No |
| 112 | Ccnd2 | 9517 | -0.267 | -0.1610 | No |
| 113 | Plxnb2 | 9699 | -0.280 | -0.1694 | No |
| 114 | Ip6k2 | 9798 | -0.288 | -0.1709 | No |
| 115 | Fdxr | 9818 | -0.290 | -0.1660 | No |
| 116 | Rpl18 | 9865 | -0.294 | -0.1631 | No |
| 117 | S100a10 | 9896 | -0.297 | -0.1589 | No |
| 118 | Btg2 | 9904 | -0.298 | -0.1528 | No |
| 119 | Sphk1 | 10082 | -0.314 | -0.1602 | No |
| 120 | Pitpnc1 | 10147 | -0.320 | -0.1582 | No |
| 121 | H2aj | 10156 | -0.321 | -0.1517 | No |
| 122 | Fam162a | 10236 | -0.329 | -0.1507 | No |
| 123 | Nol8 | 10459 | -0.353 | -0.1609 | No |
| 124 | Blcap | 10625 | -0.371 | -0.1660 | No |
| 125 | Tspyl2 | 10693 | -0.379 | -0.1629 | No |
| 126 | Cebpa | 10724 | -0.384 | -0.1568 | No |
| 127 | Def6 | 10767 | -0.388 | -0.1515 | No |
| 128 | Tprkb | 10882 | -0.401 | -0.1518 | No |
| 129 | Ei24 | 10909 | -0.406 | -0.1448 | No |
| 130 | Cd81 | 10933 | -0.408 | -0.1376 | No |
| 131 | Steap3 | 10957 | -0.412 | -0.1302 | No |
| 132 | Prkab1 | 11006 | -0.417 | -0.1248 | No |
| 133 | Ddit3 | 11137 | -0.437 | -0.1255 | No |
| 134 | Sp1 | 11370 | -0.470 | -0.1339 | No |
| 135 | Cdkn2aip | 11412 | -0.477 | -0.1266 | No |
| 136 | Bax | 11415 | -0.477 | -0.1160 | No |
| 137 | Mknk2 | 11488 | -0.489 | -0.1109 | No |
| 138 | Abhd4 | 11594 | -0.509 | -0.1081 | No |
| 139 | Irak1 | 11888 | -0.584 | -0.1188 | No |
| 140 | Gm2a | 11941 | -0.603 | -0.1095 | No |
| 141 | Sertad3 | 12001 | -0.620 | -0.1004 | No |
| 142 | Rps27l | 12015 | -0.624 | -0.0875 | No |
| 143 | Irag2 | 12069 | -0.642 | -0.0775 | No |
| 144 | Gadd45a | 12109 | -0.660 | -0.0659 | No |
| 145 | Ndrg1 | 12192 | -0.705 | -0.0568 | No |
| 146 | Tgfb1 | 12207 | -0.714 | -0.0419 | No |
| 147 | Rack1 | 12234 | -0.727 | -0.0277 | No |
| 148 | Baiap2 | 12312 | -0.812 | -0.0158 | No |
| 149 | Rpl36 | 12428 | -1.171 | 0.0011 | No |