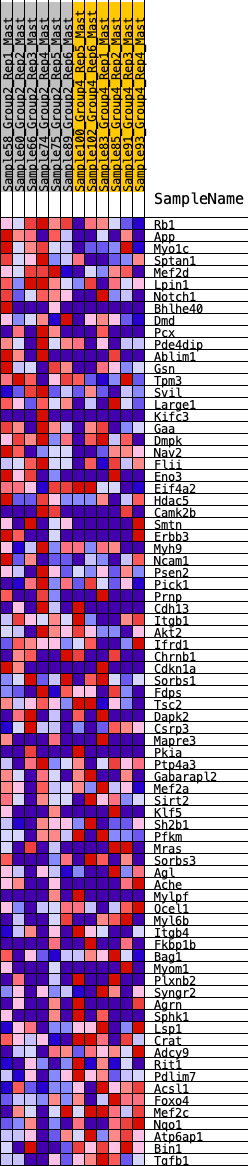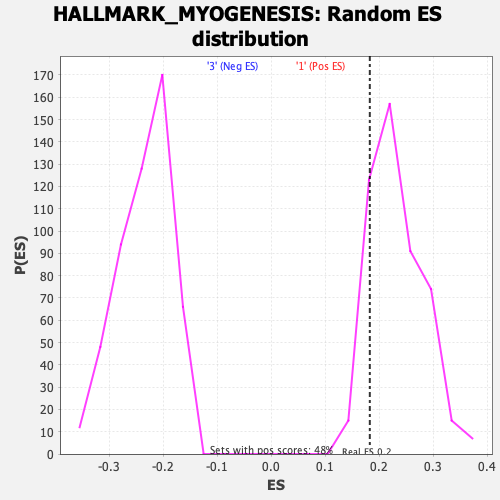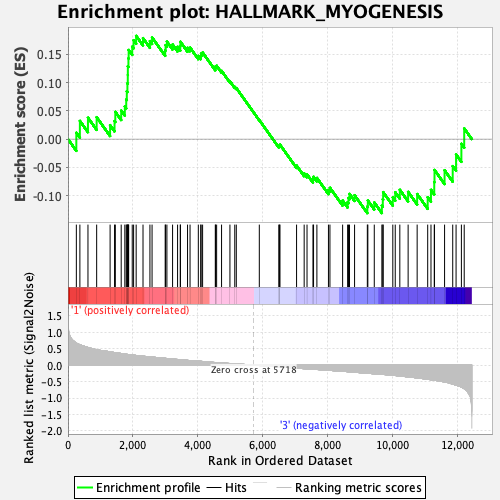
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.18269503 |
| Normalized Enrichment Score (NES) | 0.7902553 |
| Nominal p-value | 0.8443983 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rb1 | 257 | 0.670 | 0.0113 | Yes |
| 2 | App | 366 | 0.620 | 0.0322 | Yes |
| 3 | Myo1c | 615 | 0.537 | 0.0379 | Yes |
| 4 | Sptan1 | 883 | 0.469 | 0.0387 | Yes |
| 5 | Mef2d | 1297 | 0.403 | 0.0246 | Yes |
| 6 | Lpin1 | 1434 | 0.382 | 0.0319 | Yes |
| 7 | Notch1 | 1458 | 0.379 | 0.0482 | Yes |
| 8 | Bhlhe40 | 1639 | 0.352 | 0.0506 | Yes |
| 9 | Dmd | 1751 | 0.337 | 0.0577 | Yes |
| 10 | Pcx | 1794 | 0.331 | 0.0702 | Yes |
| 11 | Pde4dip | 1812 | 0.329 | 0.0845 | Yes |
| 12 | Ablim1 | 1831 | 0.325 | 0.0987 | Yes |
| 13 | Gsn | 1842 | 0.325 | 0.1134 | Yes |
| 14 | Tpm3 | 1844 | 0.325 | 0.1289 | Yes |
| 15 | Svil | 1862 | 0.322 | 0.1429 | Yes |
| 16 | Large1 | 1869 | 0.321 | 0.1578 | Yes |
| 17 | Kifc3 | 1979 | 0.309 | 0.1637 | Yes |
| 18 | Gaa | 2021 | 0.304 | 0.1750 | Yes |
| 19 | Dmpk | 2100 | 0.293 | 0.1827 | Yes |
| 20 | Nav2 | 2311 | 0.271 | 0.1787 | No |
| 21 | Flii | 2524 | 0.250 | 0.1735 | No |
| 22 | Eno3 | 2590 | 0.244 | 0.1799 | No |
| 23 | Eif4a2 | 2994 | 0.208 | 0.1572 | No |
| 24 | Hdac5 | 3006 | 0.206 | 0.1662 | No |
| 25 | Camk2b | 3047 | 0.201 | 0.1726 | No |
| 26 | Smtn | 3222 | 0.185 | 0.1674 | No |
| 27 | Erbb3 | 3376 | 0.173 | 0.1633 | No |
| 28 | Myh9 | 3460 | 0.166 | 0.1645 | No |
| 29 | Ncam1 | 3463 | 0.165 | 0.1723 | No |
| 30 | Psen2 | 3683 | 0.145 | 0.1615 | No |
| 31 | Pick1 | 3760 | 0.138 | 0.1620 | No |
| 32 | Prnp | 4015 | 0.121 | 0.1473 | No |
| 33 | Cdh13 | 4089 | 0.116 | 0.1469 | No |
| 34 | Itgb1 | 4101 | 0.115 | 0.1516 | No |
| 35 | Akt2 | 4147 | 0.111 | 0.1533 | No |
| 36 | Ifrd1 | 4538 | 0.080 | 0.1255 | No |
| 37 | Chrnb1 | 4546 | 0.079 | 0.1288 | No |
| 38 | Cdkn1a | 4579 | 0.077 | 0.1298 | No |
| 39 | Sorbs1 | 4731 | 0.066 | 0.1208 | No |
| 40 | Fdps | 4989 | 0.050 | 0.1024 | No |
| 41 | Tsc2 | 5138 | 0.040 | 0.0923 | No |
| 42 | Dapk2 | 5185 | 0.037 | 0.0904 | No |
| 43 | Csrp3 | 5894 | -0.011 | 0.0337 | No |
| 44 | Mapre3 | 6494 | -0.053 | -0.0123 | No |
| 45 | Pkia | 6519 | -0.054 | -0.0116 | No |
| 46 | Ptp4a3 | 6529 | -0.055 | -0.0097 | No |
| 47 | Gabarapl2 | 7042 | -0.090 | -0.0468 | No |
| 48 | Mef2a | 7276 | -0.106 | -0.0606 | No |
| 49 | Sirt2 | 7364 | -0.111 | -0.0623 | No |
| 50 | Klf5 | 7547 | -0.122 | -0.0712 | No |
| 51 | Sh2b1 | 7565 | -0.123 | -0.0667 | No |
| 52 | Pfkm | 7668 | -0.131 | -0.0687 | No |
| 53 | Mras | 8024 | -0.154 | -0.0900 | No |
| 54 | Sorbs3 | 8070 | -0.158 | -0.0861 | No |
| 55 | Agl | 8461 | -0.186 | -0.1087 | No |
| 56 | Ache | 8615 | -0.197 | -0.1117 | No |
| 57 | Mylpf | 8641 | -0.199 | -0.1042 | No |
| 58 | Ocel1 | 8673 | -0.202 | -0.0970 | No |
| 59 | Myl6b | 8829 | -0.213 | -0.0993 | No |
| 60 | Itgb4 | 9222 | -0.246 | -0.1193 | No |
| 61 | Fkbp1b | 9238 | -0.247 | -0.1087 | No |
| 62 | Bag1 | 9436 | -0.261 | -0.1121 | No |
| 63 | Myom1 | 9670 | -0.278 | -0.1177 | No |
| 64 | Plxnb2 | 9699 | -0.280 | -0.1065 | No |
| 65 | Syngr2 | 9710 | -0.281 | -0.0939 | No |
| 66 | Agrn | 10007 | -0.307 | -0.1031 | No |
| 67 | Sphk1 | 10082 | -0.314 | -0.0941 | No |
| 68 | Lsp1 | 10223 | -0.328 | -0.0897 | No |
| 69 | Crat | 10479 | -0.354 | -0.0933 | No |
| 70 | Adcy9 | 10757 | -0.388 | -0.0972 | No |
| 71 | Rit1 | 11082 | -0.430 | -0.1028 | No |
| 72 | Pdlim7 | 11183 | -0.442 | -0.0897 | No |
| 73 | Acsl1 | 11284 | -0.457 | -0.0760 | No |
| 74 | Foxo4 | 11292 | -0.458 | -0.0546 | No |
| 75 | Mef2c | 11602 | -0.511 | -0.0551 | No |
| 76 | Nqo1 | 11853 | -0.575 | -0.0478 | No |
| 77 | Atp6ap1 | 11956 | -0.606 | -0.0270 | No |
| 78 | Bin1 | 12120 | -0.667 | -0.0083 | No |
| 79 | Tgfb1 | 12207 | -0.714 | 0.0189 | No |