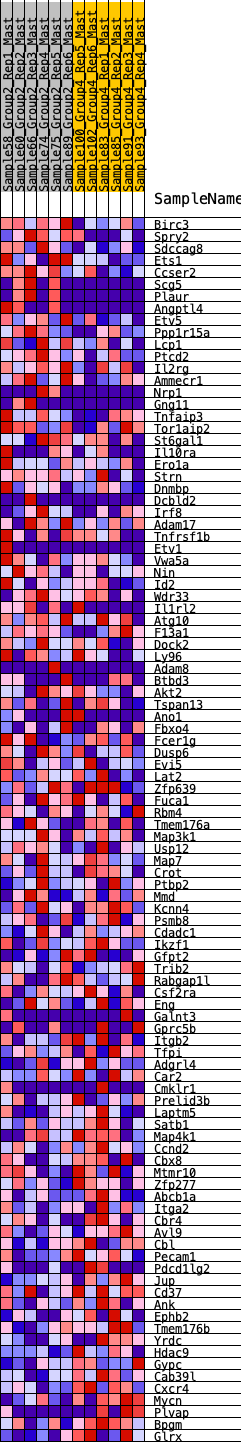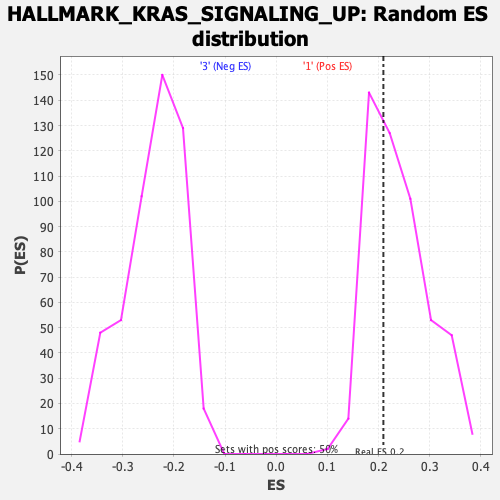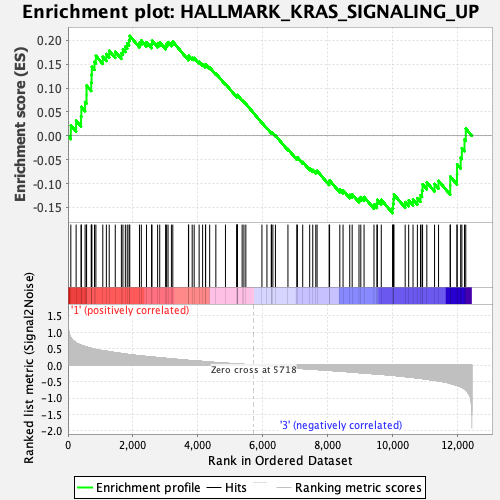
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.20937376 |
| Normalized Enrichment Score (NES) | 0.87770987 |
| Nominal p-value | 0.62828285 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Birc3 | 87 | 0.835 | 0.0217 | Yes |
| 2 | Spry2 | 250 | 0.676 | 0.0319 | Yes |
| 3 | Sdccag8 | 402 | 0.607 | 0.0406 | Yes |
| 4 | Ets1 | 412 | 0.604 | 0.0607 | Yes |
| 5 | Ccser2 | 527 | 0.563 | 0.0708 | Yes |
| 6 | Scg5 | 571 | 0.550 | 0.0863 | Yes |
| 7 | Plaur | 572 | 0.550 | 0.1053 | Yes |
| 8 | Angptl4 | 715 | 0.509 | 0.1113 | Yes |
| 9 | Etv5 | 726 | 0.506 | 0.1280 | Yes |
| 10 | Ppp1r15a | 735 | 0.502 | 0.1446 | Yes |
| 11 | Lcp1 | 816 | 0.482 | 0.1548 | Yes |
| 12 | Ptcd2 | 859 | 0.474 | 0.1677 | Yes |
| 13 | Il2rg | 1072 | 0.436 | 0.1655 | Yes |
| 14 | Ammecr1 | 1182 | 0.423 | 0.1713 | Yes |
| 15 | Nrp1 | 1270 | 0.406 | 0.1783 | Yes |
| 16 | Gng11 | 1456 | 0.379 | 0.1764 | Yes |
| 17 | Tnfaip3 | 1645 | 0.352 | 0.1732 | Yes |
| 18 | Tor1aip2 | 1692 | 0.347 | 0.1815 | Yes |
| 19 | St6gal1 | 1767 | 0.335 | 0.1870 | Yes |
| 20 | Il10ra | 1825 | 0.326 | 0.1936 | Yes |
| 21 | Ero1a | 1881 | 0.319 | 0.2002 | Yes |
| 22 | Strn | 1903 | 0.316 | 0.2094 | Yes |
| 23 | Dnmbp | 2202 | 0.281 | 0.1949 | No |
| 24 | Dcbld2 | 2259 | 0.276 | 0.1999 | No |
| 25 | Irf8 | 2418 | 0.261 | 0.1961 | No |
| 26 | Adam17 | 2573 | 0.246 | 0.1920 | No |
| 27 | Tnfrsf1b | 2588 | 0.244 | 0.1993 | No |
| 28 | Etv1 | 2761 | 0.229 | 0.1933 | No |
| 29 | Vwa5a | 2826 | 0.224 | 0.1958 | No |
| 30 | Nin | 3007 | 0.206 | 0.1883 | No |
| 31 | Id2 | 3038 | 0.203 | 0.1929 | No |
| 32 | Wdr33 | 3082 | 0.198 | 0.1962 | No |
| 33 | Il1rl2 | 3186 | 0.189 | 0.1944 | No |
| 34 | Atg10 | 3229 | 0.185 | 0.1973 | No |
| 35 | F13a1 | 3711 | 0.142 | 0.1633 | No |
| 36 | Dock2 | 3717 | 0.142 | 0.1677 | No |
| 37 | Ly96 | 3826 | 0.134 | 0.1636 | No |
| 38 | Adam8 | 3887 | 0.130 | 0.1632 | No |
| 39 | Btbd3 | 4040 | 0.120 | 0.1551 | No |
| 40 | Akt2 | 4147 | 0.111 | 0.1503 | No |
| 41 | Tspan13 | 4235 | 0.104 | 0.1468 | No |
| 42 | Ano1 | 4241 | 0.104 | 0.1500 | No |
| 43 | Fbxo4 | 4365 | 0.093 | 0.1432 | No |
| 44 | Fcer1g | 4555 | 0.079 | 0.1306 | No |
| 45 | Dusp6 | 4851 | 0.058 | 0.1087 | No |
| 46 | Evi5 | 5200 | 0.036 | 0.0818 | No |
| 47 | Lat2 | 5210 | 0.036 | 0.0823 | No |
| 48 | Zfp639 | 5213 | 0.035 | 0.0833 | No |
| 49 | Fuca1 | 5216 | 0.035 | 0.0844 | No |
| 50 | Rbm4 | 5218 | 0.035 | 0.0855 | No |
| 51 | Tmem176a | 5365 | 0.027 | 0.0746 | No |
| 52 | Map3k1 | 5420 | 0.022 | 0.0710 | No |
| 53 | Usp12 | 5479 | 0.018 | 0.0669 | No |
| 54 | Map7 | 5975 | -0.016 | 0.0274 | No |
| 55 | Crot | 6129 | -0.028 | 0.0160 | No |
| 56 | Ptbp2 | 6263 | -0.038 | 0.0065 | No |
| 57 | Mmd | 6269 | -0.038 | 0.0074 | No |
| 58 | Kcnn4 | 6312 | -0.041 | 0.0054 | No |
| 59 | Psmb8 | 6391 | -0.047 | 0.0007 | No |
| 60 | Cdadc1 | 6776 | -0.072 | -0.0280 | No |
| 61 | Ikzf1 | 7049 | -0.091 | -0.0469 | No |
| 62 | Gfpt2 | 7067 | -0.091 | -0.0451 | No |
| 63 | Trib2 | 7231 | -0.103 | -0.0548 | No |
| 64 | Rabgap1l | 7446 | -0.116 | -0.0681 | No |
| 65 | Csf2ra | 7541 | -0.121 | -0.0715 | No |
| 66 | Eng | 7634 | -0.129 | -0.0745 | No |
| 67 | Galnt3 | 7674 | -0.131 | -0.0732 | No |
| 68 | Gprc5b | 8045 | -0.156 | -0.0978 | No |
| 69 | Itgb2 | 8059 | -0.157 | -0.0934 | No |
| 70 | Tfpi | 8374 | -0.181 | -0.1126 | No |
| 71 | Adgrl4 | 8474 | -0.187 | -0.1142 | No |
| 72 | Car2 | 8680 | -0.202 | -0.1239 | No |
| 73 | Cmklr1 | 8755 | -0.207 | -0.1227 | No |
| 74 | Prelid3b | 8965 | -0.225 | -0.1319 | No |
| 75 | Laptm5 | 9021 | -0.228 | -0.1285 | No |
| 76 | Satb1 | 9121 | -0.238 | -0.1283 | No |
| 77 | Map4k1 | 9427 | -0.260 | -0.1441 | No |
| 78 | Ccnd2 | 9517 | -0.267 | -0.1421 | No |
| 79 | Cbx8 | 9532 | -0.268 | -0.1340 | No |
| 80 | Mtmr10 | 9652 | -0.276 | -0.1341 | No |
| 81 | Zfp277 | 9999 | -0.306 | -0.1516 | No |
| 82 | Abcb1a | 10016 | -0.307 | -0.1423 | No |
| 83 | Itga2 | 10030 | -0.309 | -0.1327 | No |
| 84 | Cbr4 | 10042 | -0.310 | -0.1229 | No |
| 85 | Avl9 | 10388 | -0.346 | -0.1389 | No |
| 86 | Cbl | 10496 | -0.356 | -0.1353 | No |
| 87 | Pecam1 | 10630 | -0.372 | -0.1333 | No |
| 88 | Pdcd1lg2 | 10764 | -0.388 | -0.1307 | No |
| 89 | Jup | 10860 | -0.399 | -0.1246 | No |
| 90 | Cd37 | 10907 | -0.405 | -0.1144 | No |
| 91 | Ank | 10924 | -0.407 | -0.1016 | No |
| 92 | Ephb2 | 11055 | -0.426 | -0.0975 | No |
| 93 | Tmem176b | 11293 | -0.458 | -0.1009 | No |
| 94 | Yrdc | 11416 | -0.477 | -0.0943 | No |
| 95 | Hdac9 | 11776 | -0.553 | -0.1044 | No |
| 96 | Gypc | 11778 | -0.554 | -0.0854 | No |
| 97 | Cab39l | 11985 | -0.615 | -0.0809 | No |
| 98 | Cxcr4 | 11988 | -0.617 | -0.0597 | No |
| 99 | Mycn | 12098 | -0.652 | -0.0461 | No |
| 100 | Plvap | 12135 | -0.672 | -0.0259 | No |
| 101 | Bpgm | 12216 | -0.718 | -0.0076 | No |
| 102 | Glrx | 12255 | -0.747 | 0.0151 | No |