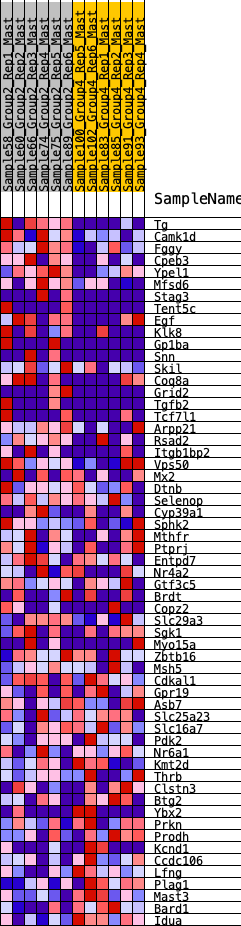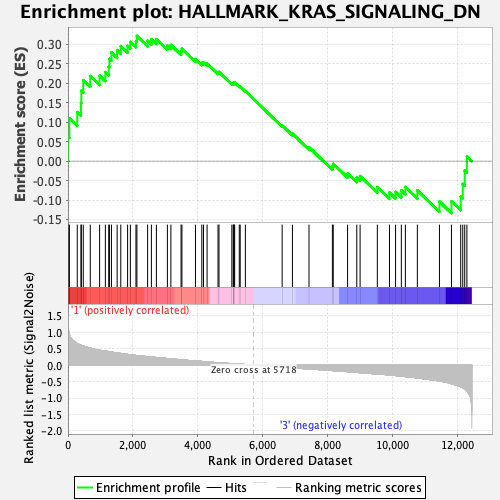
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.3218001 |
| Normalized Enrichment Score (NES) | 1.166061 |
| Nominal p-value | 0.24848485 |
| FDR q-value | 0.9812432 |
| FWER p-Value | 0.936 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tg | 9 | 1.164 | 0.0622 | Yes |
| 2 | Camk1d | 43 | 0.934 | 0.1101 | Yes |
| 3 | Fggy | 285 | 0.653 | 0.1259 | Yes |
| 4 | Cpeb3 | 400 | 0.607 | 0.1495 | Yes |
| 5 | Ypel1 | 411 | 0.604 | 0.1814 | Yes |
| 6 | Mfsd6 | 471 | 0.581 | 0.2080 | Yes |
| 7 | Stag3 | 686 | 0.515 | 0.2186 | Yes |
| 8 | Tent5c | 974 | 0.451 | 0.2197 | Yes |
| 9 | Egf | 1151 | 0.427 | 0.2286 | Yes |
| 10 | Klk8 | 1255 | 0.409 | 0.2424 | Yes |
| 11 | Gp1ba | 1276 | 0.406 | 0.2627 | Yes |
| 12 | Snn | 1338 | 0.397 | 0.2793 | Yes |
| 13 | Skil | 1514 | 0.372 | 0.2852 | Yes |
| 14 | Coq8a | 1626 | 0.355 | 0.2954 | Yes |
| 15 | Grid2 | 1836 | 0.325 | 0.2961 | Yes |
| 16 | Tgfb2 | 1921 | 0.314 | 0.3063 | Yes |
| 17 | Tcf7l1 | 2094 | 0.294 | 0.3083 | Yes |
| 18 | Arpp21 | 2123 | 0.291 | 0.3218 | Yes |
| 19 | Rsad2 | 2453 | 0.257 | 0.3091 | No |
| 20 | Itgb1bp2 | 2569 | 0.246 | 0.3132 | No |
| 21 | Vps50 | 2723 | 0.232 | 0.3133 | No |
| 22 | Mx2 | 3062 | 0.200 | 0.2968 | No |
| 23 | Dtnb | 3170 | 0.190 | 0.2985 | No |
| 24 | Selenop | 3483 | 0.163 | 0.2821 | No |
| 25 | Cyp39a1 | 3510 | 0.161 | 0.2887 | No |
| 26 | Sphk2 | 3927 | 0.129 | 0.2621 | No |
| 27 | Mthfr | 4123 | 0.113 | 0.2524 | No |
| 28 | Ptprj | 4174 | 0.109 | 0.2543 | No |
| 29 | Entpd7 | 4285 | 0.099 | 0.2508 | No |
| 30 | Nr4a2 | 4622 | 0.072 | 0.2276 | No |
| 31 | Gtf3c5 | 4652 | 0.070 | 0.2290 | No |
| 32 | Brdt | 5048 | 0.046 | 0.1996 | No |
| 33 | Copz2 | 5091 | 0.043 | 0.1985 | No |
| 34 | Slc29a3 | 5108 | 0.042 | 0.1995 | No |
| 35 | Sgk1 | 5112 | 0.042 | 0.2015 | No |
| 36 | Myo15a | 5135 | 0.040 | 0.2019 | No |
| 37 | Zbtb16 | 5277 | 0.031 | 0.1923 | No |
| 38 | Msh5 | 5305 | 0.030 | 0.1917 | No |
| 39 | Cdkal1 | 5465 | 0.019 | 0.1799 | No |
| 40 | Gpr19 | 6598 | -0.060 | 0.0917 | No |
| 41 | Asb7 | 6914 | -0.081 | 0.0706 | No |
| 42 | Slc25a23 | 7425 | -0.115 | 0.0357 | No |
| 43 | Slc16a7 | 8145 | -0.164 | -0.0135 | No |
| 44 | Pdk2 | 8172 | -0.165 | -0.0067 | No |
| 45 | Nr6a1 | 8614 | -0.197 | -0.0317 | No |
| 46 | Kmt2d | 8897 | -0.219 | -0.0426 | No |
| 47 | Thrb | 8999 | -0.227 | -0.0385 | No |
| 48 | Clstn3 | 9530 | -0.268 | -0.0668 | No |
| 49 | Btg2 | 9904 | -0.298 | -0.0808 | No |
| 50 | Ybx2 | 10092 | -0.315 | -0.0789 | No |
| 51 | Prkn | 10270 | -0.333 | -0.0752 | No |
| 52 | Prodh | 10394 | -0.347 | -0.0664 | No |
| 53 | Kcnd1 | 10763 | -0.388 | -0.0751 | No |
| 54 | Ccdc106 | 11444 | -0.482 | -0.1039 | No |
| 55 | Lfng | 11814 | -0.563 | -0.1033 | No |
| 56 | Plag1 | 12099 | -0.653 | -0.0909 | No |
| 57 | Mast3 | 12162 | -0.689 | -0.0587 | No |
| 58 | Bard1 | 12223 | -0.721 | -0.0246 | No |
| 59 | Idua | 12290 | -0.779 | 0.0122 | No |