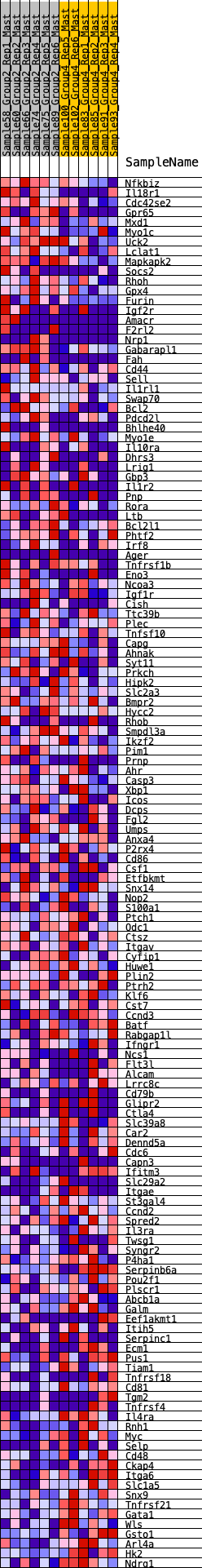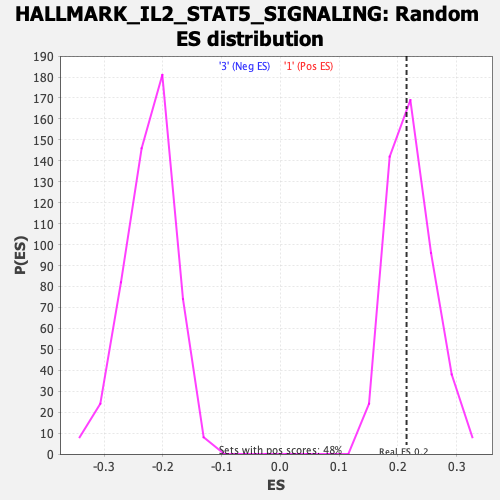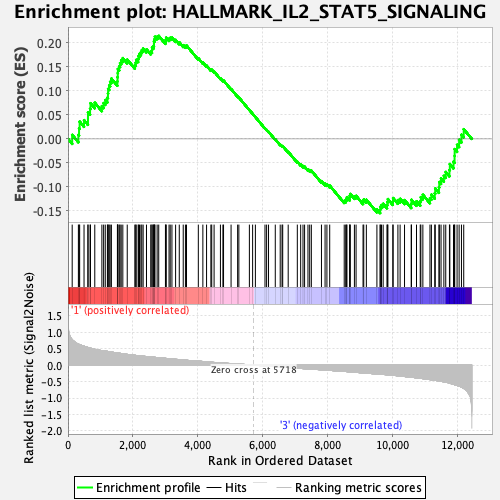
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | 0.2146275 |
| Normalized Enrichment Score (NES) | 0.9704743 |
| Nominal p-value | 0.5324948 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfkbiz | 129 | 0.776 | 0.0084 | Yes |
| 2 | Il18r1 | 320 | 0.638 | 0.0085 | Yes |
| 3 | Cdc42se2 | 341 | 0.630 | 0.0222 | Yes |
| 4 | Gpr65 | 359 | 0.623 | 0.0360 | Yes |
| 5 | Mxd1 | 494 | 0.574 | 0.0390 | Yes |
| 6 | Myo1c | 615 | 0.537 | 0.0423 | Yes |
| 7 | Uck2 | 616 | 0.537 | 0.0554 | Yes |
| 8 | Lclat1 | 680 | 0.516 | 0.0628 | Yes |
| 9 | Mapkapk2 | 693 | 0.514 | 0.0744 | Yes |
| 10 | Socs2 | 825 | 0.480 | 0.0754 | Yes |
| 11 | Rhoh | 1044 | 0.440 | 0.0684 | Yes |
| 12 | Gpx4 | 1098 | 0.434 | 0.0746 | Yes |
| 13 | Furin | 1148 | 0.427 | 0.0810 | Yes |
| 14 | Igf2r | 1214 | 0.418 | 0.0859 | Yes |
| 15 | Amacr | 1231 | 0.412 | 0.0946 | Yes |
| 16 | F2rl2 | 1237 | 0.412 | 0.1043 | Yes |
| 17 | Nrp1 | 1270 | 0.406 | 0.1115 | Yes |
| 18 | Gabarapl1 | 1299 | 0.403 | 0.1191 | Yes |
| 19 | Fah | 1337 | 0.397 | 0.1257 | Yes |
| 20 | Cd44 | 1522 | 0.371 | 0.1198 | Yes |
| 21 | Sell | 1526 | 0.371 | 0.1286 | Yes |
| 22 | Il1rl1 | 1533 | 0.369 | 0.1371 | Yes |
| 23 | Swap70 | 1537 | 0.369 | 0.1458 | Yes |
| 24 | Bcl2 | 1579 | 0.362 | 0.1513 | Yes |
| 25 | Pdcd2l | 1604 | 0.359 | 0.1580 | Yes |
| 26 | Bhlhe40 | 1639 | 0.352 | 0.1638 | Yes |
| 27 | Myo1e | 1689 | 0.347 | 0.1683 | Yes |
| 28 | Il10ra | 1825 | 0.326 | 0.1653 | Yes |
| 29 | Dhrs3 | 2059 | 0.300 | 0.1536 | Yes |
| 30 | Lrig1 | 2086 | 0.295 | 0.1587 | Yes |
| 31 | Gbp3 | 2101 | 0.293 | 0.1647 | Yes |
| 32 | Il1r2 | 2161 | 0.285 | 0.1668 | Yes |
| 33 | Pnp | 2175 | 0.283 | 0.1727 | Yes |
| 34 | Rora | 2203 | 0.281 | 0.1773 | Yes |
| 35 | Ltb | 2246 | 0.277 | 0.1806 | Yes |
| 36 | Bcl2l1 | 2275 | 0.274 | 0.1850 | Yes |
| 37 | Phtf2 | 2315 | 0.270 | 0.1884 | Yes |
| 38 | Irf8 | 2418 | 0.261 | 0.1865 | Yes |
| 39 | Ager | 2549 | 0.247 | 0.1819 | Yes |
| 40 | Tnfrsf1b | 2588 | 0.244 | 0.1848 | Yes |
| 41 | Eno3 | 2590 | 0.244 | 0.1906 | Yes |
| 42 | Ncoa3 | 2630 | 0.240 | 0.1933 | Yes |
| 43 | Igf1r | 2648 | 0.239 | 0.1977 | Yes |
| 44 | Cish | 2649 | 0.239 | 0.2035 | Yes |
| 45 | Ttc39b | 2668 | 0.237 | 0.2078 | Yes |
| 46 | Plec | 2678 | 0.237 | 0.2129 | Yes |
| 47 | Tnfsf10 | 2746 | 0.230 | 0.2130 | Yes |
| 48 | Capg | 2795 | 0.226 | 0.2146 | Yes |
| 49 | Ahnak | 3002 | 0.207 | 0.2029 | No |
| 50 | Syt11 | 3014 | 0.205 | 0.2070 | No |
| 51 | Prkch | 3025 | 0.203 | 0.2111 | No |
| 52 | Hipk2 | 3113 | 0.197 | 0.2089 | No |
| 53 | Slc2a3 | 3149 | 0.192 | 0.2107 | No |
| 54 | Bmpr2 | 3202 | 0.187 | 0.2110 | No |
| 55 | Hycc2 | 3315 | 0.177 | 0.2062 | No |
| 56 | Rhob | 3428 | 0.169 | 0.2012 | No |
| 57 | Smpdl3a | 3550 | 0.159 | 0.1952 | No |
| 58 | Ikzf2 | 3618 | 0.152 | 0.1935 | No |
| 59 | Pim1 | 3651 | 0.148 | 0.1945 | No |
| 60 | Prnp | 4015 | 0.121 | 0.1679 | No |
| 61 | Ahr | 4155 | 0.111 | 0.1593 | No |
| 62 | Casp3 | 4269 | 0.101 | 0.1526 | No |
| 63 | Xbp1 | 4402 | 0.090 | 0.1441 | No |
| 64 | Icos | 4428 | 0.088 | 0.1442 | No |
| 65 | Dcps | 4501 | 0.082 | 0.1403 | No |
| 66 | Fgl2 | 4696 | 0.068 | 0.1262 | No |
| 67 | Umps | 4773 | 0.063 | 0.1216 | No |
| 68 | Anxa4 | 4792 | 0.062 | 0.1216 | No |
| 69 | P2rx4 | 5024 | 0.047 | 0.1040 | No |
| 70 | Cd86 | 5226 | 0.035 | 0.0885 | No |
| 71 | Csf1 | 5264 | 0.032 | 0.0863 | No |
| 72 | Etfbkmt | 5588 | 0.011 | 0.0603 | No |
| 73 | Snx14 | 5688 | 0.003 | 0.0523 | No |
| 74 | Nop2 | 5773 | -0.002 | 0.0455 | No |
| 75 | S100a1 | 6069 | -0.024 | 0.0221 | No |
| 76 | Ptch1 | 6119 | -0.028 | 0.0188 | No |
| 77 | Odc1 | 6178 | -0.031 | 0.0148 | No |
| 78 | Ctsz | 6386 | -0.046 | -0.0009 | No |
| 79 | Itgav | 6538 | -0.056 | -0.0118 | No |
| 80 | Cyfip1 | 6594 | -0.059 | -0.0148 | No |
| 81 | Huwe1 | 6615 | -0.061 | -0.0150 | No |
| 82 | Plin2 | 6784 | -0.073 | -0.0269 | No |
| 83 | Ptrh2 | 7065 | -0.091 | -0.0474 | No |
| 84 | Klf6 | 7168 | -0.099 | -0.0533 | No |
| 85 | Cst7 | 7240 | -0.104 | -0.0565 | No |
| 86 | Ccnd3 | 7289 | -0.106 | -0.0578 | No |
| 87 | Batf | 7393 | -0.113 | -0.0635 | No |
| 88 | Rabgap1l | 7446 | -0.116 | -0.0649 | No |
| 89 | Ifngr1 | 7497 | -0.119 | -0.0660 | No |
| 90 | Ncs1 | 7809 | -0.139 | -0.0879 | No |
| 91 | Flt3l | 7920 | -0.147 | -0.0933 | No |
| 92 | Alcam | 7977 | -0.150 | -0.0942 | No |
| 93 | Lrrc8c | 8063 | -0.157 | -0.0973 | No |
| 94 | Cd79b | 8510 | -0.189 | -0.1289 | No |
| 95 | Glipr2 | 8556 | -0.192 | -0.1279 | No |
| 96 | Ctla4 | 8576 | -0.193 | -0.1248 | No |
| 97 | Slc39a8 | 8594 | -0.195 | -0.1214 | No |
| 98 | Car2 | 8680 | -0.202 | -0.1234 | No |
| 99 | Dennd5a | 8681 | -0.202 | -0.1184 | No |
| 100 | Cdc6 | 8696 | -0.203 | -0.1146 | No |
| 101 | Capn3 | 8824 | -0.213 | -0.1198 | No |
| 102 | Ifitm3 | 8875 | -0.218 | -0.1186 | No |
| 103 | Slc29a2 | 9090 | -0.234 | -0.1303 | No |
| 104 | Itgae | 9112 | -0.236 | -0.1262 | No |
| 105 | St3gal4 | 9192 | -0.243 | -0.1267 | No |
| 106 | Ccnd2 | 9517 | -0.267 | -0.1466 | No |
| 107 | Spred2 | 9617 | -0.274 | -0.1480 | No |
| 108 | Il3ra | 9620 | -0.274 | -0.1414 | No |
| 109 | Twsg1 | 9659 | -0.277 | -0.1378 | No |
| 110 | Syngr2 | 9710 | -0.281 | -0.1350 | No |
| 111 | P4ha1 | 9829 | -0.291 | -0.1375 | No |
| 112 | Serpinb6a | 9840 | -0.292 | -0.1312 | No |
| 113 | Pou2f1 | 9856 | -0.293 | -0.1253 | No |
| 114 | Plscr1 | 10008 | -0.307 | -0.1301 | No |
| 115 | Abcb1a | 10016 | -0.307 | -0.1232 | No |
| 116 | Galm | 10164 | -0.322 | -0.1274 | No |
| 117 | Eef1akmt1 | 10232 | -0.329 | -0.1248 | No |
| 118 | Itih5 | 10368 | -0.344 | -0.1274 | No |
| 119 | Serpinc1 | 10574 | -0.366 | -0.1352 | No |
| 120 | Ecm1 | 10582 | -0.367 | -0.1268 | No |
| 121 | Pus1 | 10737 | -0.386 | -0.1300 | No |
| 122 | Tiam1 | 10852 | -0.398 | -0.1295 | No |
| 123 | Tnfrsf18 | 10871 | -0.400 | -0.1213 | No |
| 124 | Cd81 | 10933 | -0.408 | -0.1163 | No |
| 125 | Tgm2 | 11152 | -0.438 | -0.1234 | No |
| 126 | Tnfrsf4 | 11196 | -0.443 | -0.1161 | No |
| 127 | Il4ra | 11300 | -0.459 | -0.1133 | No |
| 128 | Rnh1 | 11314 | -0.461 | -0.1031 | No |
| 129 | Myc | 11429 | -0.480 | -0.1007 | No |
| 130 | Selp | 11439 | -0.482 | -0.0897 | No |
| 131 | Cd48 | 11493 | -0.490 | -0.0821 | No |
| 132 | Ckap4 | 11580 | -0.506 | -0.0768 | No |
| 133 | Itga6 | 11638 | -0.518 | -0.0688 | No |
| 134 | Slc1a5 | 11752 | -0.544 | -0.0648 | No |
| 135 | Snx9 | 11764 | -0.548 | -0.0523 | No |
| 136 | Tnfrsf21 | 11877 | -0.580 | -0.0473 | No |
| 137 | Gata1 | 11910 | -0.593 | -0.0355 | No |
| 138 | Wls | 11913 | -0.594 | -0.0212 | No |
| 139 | Gsto1 | 11987 | -0.616 | -0.0121 | No |
| 140 | Arl4a | 12050 | -0.634 | -0.0018 | No |
| 141 | Hk2 | 12122 | -0.668 | 0.0087 | No |
| 142 | Ndrg1 | 12192 | -0.705 | 0.0202 | No |