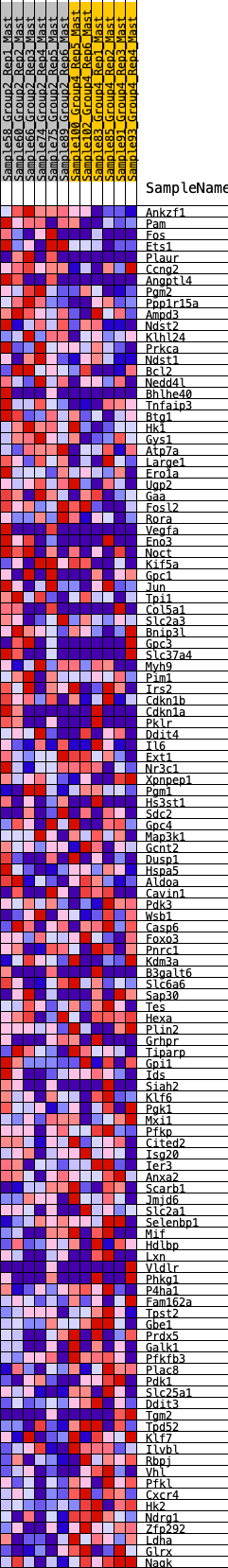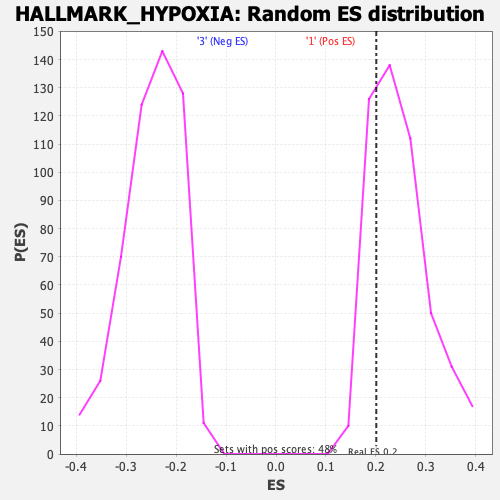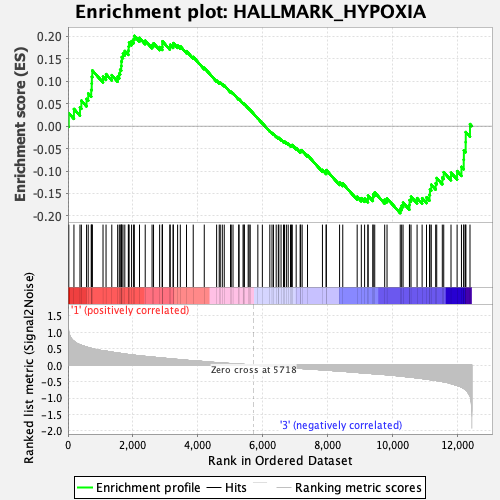
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.20105124 |
| Normalized Enrichment Score (NES) | 0.81200624 |
| Nominal p-value | 0.7871901 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ankzf1 | 24 | 1.021 | 0.0290 | Yes |
| 2 | Pam | 183 | 0.727 | 0.0382 | Yes |
| 3 | Fos | 369 | 0.619 | 0.0419 | Yes |
| 4 | Ets1 | 412 | 0.604 | 0.0568 | Yes |
| 5 | Plaur | 572 | 0.550 | 0.0606 | Yes |
| 6 | Ccng2 | 623 | 0.535 | 0.0728 | Yes |
| 7 | Angptl4 | 715 | 0.509 | 0.0808 | Yes |
| 8 | Pgm2 | 731 | 0.505 | 0.0949 | Yes |
| 9 | Ppp1r15a | 735 | 0.502 | 0.1099 | Yes |
| 10 | Ampd3 | 749 | 0.499 | 0.1240 | Yes |
| 11 | Ndst2 | 1079 | 0.435 | 0.1105 | Yes |
| 12 | Klhl24 | 1172 | 0.424 | 0.1159 | Yes |
| 13 | Prkca | 1349 | 0.396 | 0.1136 | Yes |
| 14 | Ndst1 | 1530 | 0.370 | 0.1102 | Yes |
| 15 | Bcl2 | 1579 | 0.362 | 0.1173 | Yes |
| 16 | Nedd4l | 1607 | 0.358 | 0.1259 | Yes |
| 17 | Bhlhe40 | 1639 | 0.352 | 0.1341 | Yes |
| 18 | Tnfaip3 | 1645 | 0.352 | 0.1443 | Yes |
| 19 | Btg1 | 1658 | 0.350 | 0.1540 | Yes |
| 20 | Hk1 | 1694 | 0.347 | 0.1616 | Yes |
| 21 | Gys1 | 1750 | 0.337 | 0.1674 | Yes |
| 22 | Atp7a | 1859 | 0.322 | 0.1684 | Yes |
| 23 | Large1 | 1869 | 0.321 | 0.1774 | Yes |
| 24 | Ero1a | 1881 | 0.319 | 0.1862 | Yes |
| 25 | Ugp2 | 1959 | 0.311 | 0.1893 | Yes |
| 26 | Gaa | 2021 | 0.304 | 0.1936 | Yes |
| 27 | Fosl2 | 2043 | 0.303 | 0.2011 | Yes |
| 28 | Rora | 2203 | 0.281 | 0.1967 | No |
| 29 | Vegfa | 2379 | 0.264 | 0.1904 | No |
| 30 | Eno3 | 2590 | 0.244 | 0.1808 | No |
| 31 | Noct | 2635 | 0.240 | 0.1845 | No |
| 32 | Kif5a | 2823 | 0.224 | 0.1761 | No |
| 33 | Gpc1 | 2904 | 0.216 | 0.1762 | No |
| 34 | Jun | 2908 | 0.216 | 0.1824 | No |
| 35 | Tpi1 | 2909 | 0.215 | 0.1890 | No |
| 36 | Col5a1 | 3137 | 0.194 | 0.1764 | No |
| 37 | Slc2a3 | 3149 | 0.192 | 0.1814 | No |
| 38 | Bnip3l | 3239 | 0.184 | 0.1797 | No |
| 39 | Gpc3 | 3244 | 0.183 | 0.1849 | No |
| 40 | Slc37a4 | 3377 | 0.173 | 0.1795 | No |
| 41 | Myh9 | 3460 | 0.166 | 0.1778 | No |
| 42 | Pim1 | 3651 | 0.148 | 0.1669 | No |
| 43 | Irs2 | 3858 | 0.132 | 0.1542 | No |
| 44 | Cdkn1b | 4198 | 0.107 | 0.1299 | No |
| 45 | Cdkn1a | 4579 | 0.077 | 0.1014 | No |
| 46 | Pklr | 4659 | 0.070 | 0.0971 | No |
| 47 | Ddit4 | 4690 | 0.068 | 0.0967 | No |
| 48 | Il6 | 4754 | 0.064 | 0.0936 | No |
| 49 | Ext1 | 4814 | 0.061 | 0.0906 | No |
| 50 | Nr3c1 | 5007 | 0.049 | 0.0765 | No |
| 51 | Xpnpep1 | 5029 | 0.047 | 0.0762 | No |
| 52 | Pgm1 | 5084 | 0.043 | 0.0732 | No |
| 53 | Hs3st1 | 5260 | 0.032 | 0.0600 | No |
| 54 | Sdc2 | 5269 | 0.032 | 0.0603 | No |
| 55 | Gpc4 | 5393 | 0.025 | 0.0510 | No |
| 56 | Map3k1 | 5420 | 0.022 | 0.0496 | No |
| 57 | Gcnt2 | 5434 | 0.021 | 0.0492 | No |
| 58 | Dusp1 | 5551 | 0.013 | 0.0402 | No |
| 59 | Hspa5 | 5579 | 0.012 | 0.0384 | No |
| 60 | Aldoa | 5614 | 0.009 | 0.0359 | No |
| 61 | Cavin1 | 5849 | -0.008 | 0.0171 | No |
| 62 | Pdk3 | 5988 | -0.017 | 0.0064 | No |
| 63 | Wsb1 | 6218 | -0.034 | -0.0111 | No |
| 64 | Casp6 | 6285 | -0.039 | -0.0153 | No |
| 65 | Foxo3 | 6330 | -0.042 | -0.0176 | No |
| 66 | Pnrc1 | 6414 | -0.048 | -0.0229 | No |
| 67 | Kdm3a | 6479 | -0.052 | -0.0265 | No |
| 68 | B3galt6 | 6496 | -0.053 | -0.0262 | No |
| 69 | Slc6a6 | 6564 | -0.058 | -0.0299 | No |
| 70 | Sap30 | 6639 | -0.062 | -0.0340 | No |
| 71 | Tes | 6671 | -0.064 | -0.0345 | No |
| 72 | Hexa | 6728 | -0.069 | -0.0370 | No |
| 73 | Plin2 | 6784 | -0.073 | -0.0393 | No |
| 74 | Grhpr | 6855 | -0.078 | -0.0426 | No |
| 75 | Tiparp | 6884 | -0.079 | -0.0425 | No |
| 76 | Gpi1 | 6910 | -0.081 | -0.0420 | No |
| 77 | Ids | 7030 | -0.089 | -0.0490 | No |
| 78 | Siah2 | 7152 | -0.098 | -0.0559 | No |
| 79 | Klf6 | 7168 | -0.099 | -0.0541 | No |
| 80 | Pgk1 | 7214 | -0.102 | -0.0546 | No |
| 81 | Mxi1 | 7379 | -0.112 | -0.0646 | No |
| 82 | Pfkp | 7839 | -0.141 | -0.0975 | No |
| 83 | Cited2 | 7954 | -0.148 | -0.1023 | No |
| 84 | Isg20 | 7957 | -0.148 | -0.0979 | No |
| 85 | Ier3 | 8367 | -0.180 | -0.1257 | No |
| 86 | Anxa2 | 8465 | -0.186 | -0.1279 | No |
| 87 | Scarb1 | 8909 | -0.220 | -0.1572 | No |
| 88 | Jmjd6 | 9037 | -0.230 | -0.1605 | No |
| 89 | Slc2a1 | 9138 | -0.239 | -0.1614 | No |
| 90 | Selenbp1 | 9242 | -0.247 | -0.1622 | No |
| 91 | Mif | 9243 | -0.248 | -0.1547 | No |
| 92 | Hdlbp | 9393 | -0.258 | -0.1590 | No |
| 93 | Lxn | 9401 | -0.258 | -0.1518 | No |
| 94 | Vldlr | 9450 | -0.262 | -0.1477 | No |
| 95 | Phkg1 | 9757 | -0.285 | -0.1639 | No |
| 96 | P4ha1 | 9829 | -0.291 | -0.1609 | No |
| 97 | Fam162a | 10236 | -0.329 | -0.1839 | No |
| 98 | Tpst2 | 10280 | -0.334 | -0.1772 | No |
| 99 | Gbe1 | 10323 | -0.339 | -0.1704 | No |
| 100 | Prdx5 | 10519 | -0.360 | -0.1753 | No |
| 101 | Galk1 | 10525 | -0.360 | -0.1648 | No |
| 102 | Pfkfb3 | 10568 | -0.366 | -0.1571 | No |
| 103 | Plac8 | 10755 | -0.388 | -0.1604 | No |
| 104 | Pdk1 | 10910 | -0.406 | -0.1606 | No |
| 105 | Slc25a1 | 11045 | -0.425 | -0.1586 | No |
| 106 | Ddit3 | 11137 | -0.437 | -0.1528 | No |
| 107 | Tgm2 | 11152 | -0.438 | -0.1406 | No |
| 108 | Tpd52 | 11192 | -0.443 | -0.1304 | No |
| 109 | Klf7 | 11328 | -0.464 | -0.1273 | No |
| 110 | Ilvbl | 11360 | -0.468 | -0.1156 | No |
| 111 | Rbpj | 11532 | -0.497 | -0.1144 | No |
| 112 | Vhl | 11576 | -0.505 | -0.1026 | No |
| 113 | Pfkl | 11800 | -0.560 | -0.1037 | No |
| 114 | Cxcr4 | 11988 | -0.617 | -0.1002 | No |
| 115 | Hk2 | 12122 | -0.668 | -0.0907 | No |
| 116 | Ndrg1 | 12192 | -0.705 | -0.0750 | No |
| 117 | Zfp292 | 12197 | -0.707 | -0.0539 | No |
| 118 | Ldha | 12252 | -0.745 | -0.0357 | No |
| 119 | Glrx | 12255 | -0.747 | -0.0132 | No |
| 120 | Nagk | 12385 | -0.930 | 0.0045 | No |