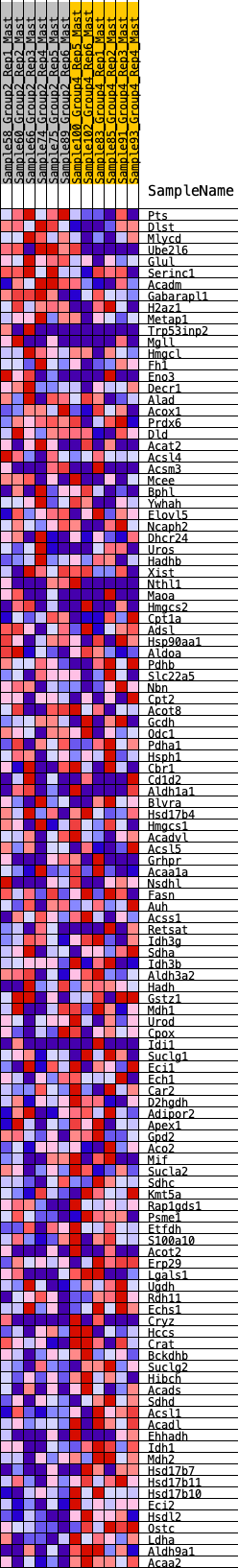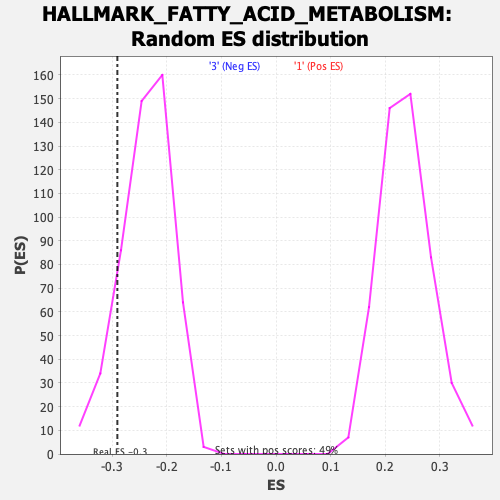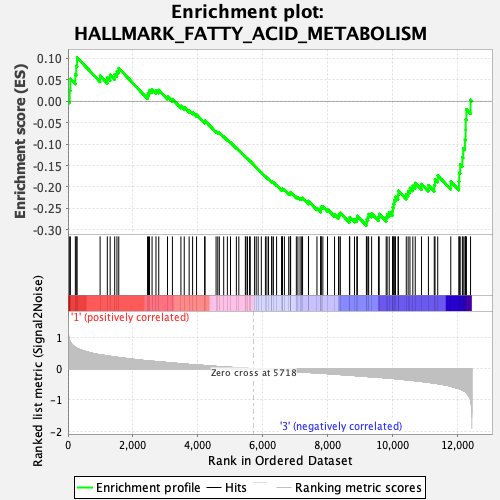
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.2902745 |
| Normalized Enrichment Score (NES) | -1.2260692 |
| Nominal p-value | 0.13188976 |
| FDR q-value | 0.66351134 |
| FWER p-Value | 0.871 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pts | 52 | 0.905 | 0.0256 | No |
| 2 | Dlst | 72 | 0.857 | 0.0524 | No |
| 3 | Mlycd | 226 | 0.690 | 0.0627 | No |
| 4 | Ube2l6 | 255 | 0.672 | 0.0826 | No |
| 5 | Glul | 281 | 0.655 | 0.1022 | No |
| 6 | Serinc1 | 989 | 0.448 | 0.0596 | No |
| 7 | Acadm | 1211 | 0.419 | 0.0555 | No |
| 8 | Gabarapl1 | 1299 | 0.403 | 0.0617 | No |
| 9 | H2az1 | 1439 | 0.381 | 0.0630 | No |
| 10 | Metap1 | 1508 | 0.372 | 0.0698 | No |
| 11 | Trp53inp2 | 1564 | 0.364 | 0.0773 | No |
| 12 | Mgll | 2451 | 0.258 | 0.0139 | No |
| 13 | Hmgcl | 2480 | 0.255 | 0.0201 | No |
| 14 | Fh1 | 2512 | 0.250 | 0.0258 | No |
| 15 | Eno3 | 2590 | 0.244 | 0.0276 | No |
| 16 | Decr1 | 2710 | 0.233 | 0.0257 | No |
| 17 | Alad | 2796 | 0.226 | 0.0262 | No |
| 18 | Acox1 | 3065 | 0.199 | 0.0110 | No |
| 19 | Prdx6 | 3217 | 0.186 | 0.0049 | No |
| 20 | Dld | 3479 | 0.164 | -0.0109 | No |
| 21 | Acat2 | 3581 | 0.155 | -0.0139 | No |
| 22 | Acsl4 | 3732 | 0.140 | -0.0215 | No |
| 23 | Acsm3 | 3839 | 0.133 | -0.0257 | No |
| 24 | Mcee | 3961 | 0.126 | -0.0314 | No |
| 25 | Bphl | 4210 | 0.106 | -0.0480 | No |
| 26 | Ywhah | 4222 | 0.105 | -0.0454 | No |
| 27 | Elovl5 | 4562 | 0.078 | -0.0703 | No |
| 28 | Ncaph2 | 4615 | 0.073 | -0.0722 | No |
| 29 | Dhcr24 | 4662 | 0.070 | -0.0736 | No |
| 30 | Uros | 4797 | 0.062 | -0.0824 | No |
| 31 | Hadhb | 4910 | 0.054 | -0.0897 | No |
| 32 | Xist | 5006 | 0.049 | -0.0958 | No |
| 33 | Nthl1 | 5187 | 0.037 | -0.1092 | No |
| 34 | Maoa | 5263 | 0.032 | -0.1142 | No |
| 35 | Hmgcs2 | 5464 | 0.019 | -0.1298 | No |
| 36 | Cpt1a | 5484 | 0.018 | -0.1308 | No |
| 37 | Adsl | 5531 | 0.014 | -0.1340 | No |
| 38 | Hsp90aa1 | 5594 | 0.010 | -0.1387 | No |
| 39 | Aldoa | 5614 | 0.009 | -0.1400 | No |
| 40 | Pdhb | 5749 | -0.001 | -0.1508 | No |
| 41 | Slc22a5 | 5799 | -0.004 | -0.1547 | No |
| 42 | Nbn | 5859 | -0.009 | -0.1592 | No |
| 43 | Cpt2 | 5956 | -0.015 | -0.1665 | No |
| 44 | Acot8 | 6082 | -0.025 | -0.1758 | No |
| 45 | Gcdh | 6128 | -0.028 | -0.1785 | No |
| 46 | Odc1 | 6178 | -0.031 | -0.1814 | No |
| 47 | Pdha1 | 6274 | -0.038 | -0.1879 | No |
| 48 | Hsph1 | 6281 | -0.039 | -0.1871 | No |
| 49 | Cbr1 | 6328 | -0.042 | -0.1895 | No |
| 50 | Cd1d2 | 6427 | -0.049 | -0.1958 | No |
| 51 | Aldh1a1 | 6578 | -0.058 | -0.2061 | No |
| 52 | Blvra | 6601 | -0.060 | -0.2059 | No |
| 53 | Hsd17b4 | 6606 | -0.060 | -0.2042 | No |
| 54 | Hmgcs1 | 6667 | -0.064 | -0.2070 | No |
| 55 | Acadvl | 6800 | -0.074 | -0.2152 | No |
| 56 | Acsl5 | 6846 | -0.077 | -0.2163 | No |
| 57 | Grhpr | 6855 | -0.078 | -0.2144 | No |
| 58 | Acaa1a | 6859 | -0.078 | -0.2121 | No |
| 59 | Nsdhl | 7035 | -0.089 | -0.2234 | No |
| 60 | Fasn | 7083 | -0.093 | -0.2241 | No |
| 61 | Auh | 7150 | -0.098 | -0.2262 | No |
| 62 | Acss1 | 7199 | -0.101 | -0.2268 | No |
| 63 | Retsat | 7227 | -0.103 | -0.2256 | No |
| 64 | Idh3g | 7408 | -0.114 | -0.2365 | No |
| 65 | Sdha | 7411 | -0.114 | -0.2329 | No |
| 66 | Idh3b | 7670 | -0.131 | -0.2495 | No |
| 67 | Aldh3a2 | 7788 | -0.138 | -0.2544 | No |
| 68 | Hadh | 7789 | -0.138 | -0.2499 | No |
| 69 | Gstz1 | 7801 | -0.139 | -0.2462 | No |
| 70 | Mdh1 | 7852 | -0.142 | -0.2455 | No |
| 71 | Urod | 7995 | -0.151 | -0.2521 | No |
| 72 | Cpox | 8212 | -0.169 | -0.2640 | No |
| 73 | Idi1 | 8335 | -0.178 | -0.2681 | No |
| 74 | Suclg1 | 8348 | -0.179 | -0.2631 | No |
| 75 | Eci1 | 8391 | -0.182 | -0.2606 | No |
| 76 | Ech1 | 8671 | -0.202 | -0.2766 | No |
| 77 | Car2 | 8680 | -0.202 | -0.2705 | No |
| 78 | D2hgdh | 8826 | -0.213 | -0.2753 | No |
| 79 | Adipor2 | 8899 | -0.219 | -0.2739 | No |
| 80 | Apex1 | 8915 | -0.221 | -0.2678 | No |
| 81 | Gpd2 | 9193 | -0.243 | -0.2823 | Yes |
| 82 | Aco2 | 9206 | -0.244 | -0.2752 | Yes |
| 83 | Mif | 9243 | -0.248 | -0.2699 | Yes |
| 84 | Sucla2 | 9263 | -0.249 | -0.2633 | Yes |
| 85 | Sdhc | 9354 | -0.255 | -0.2622 | Yes |
| 86 | Kmt5a | 9565 | -0.270 | -0.2703 | Yes |
| 87 | Rap1gds1 | 9588 | -0.272 | -0.2631 | Yes |
| 88 | Psme1 | 9803 | -0.288 | -0.2709 | Yes |
| 89 | Etfdh | 9831 | -0.291 | -0.2635 | Yes |
| 90 | S100a10 | 9896 | -0.297 | -0.2589 | Yes |
| 91 | Acot2 | 9997 | -0.306 | -0.2570 | Yes |
| 92 | Erp29 | 10005 | -0.307 | -0.2474 | Yes |
| 93 | Lgals1 | 10033 | -0.309 | -0.2394 | Yes |
| 94 | Ugdh | 10054 | -0.311 | -0.2308 | Yes |
| 95 | Rdh11 | 10089 | -0.314 | -0.2232 | Yes |
| 96 | Echs1 | 10171 | -0.322 | -0.2191 | Yes |
| 97 | Cryz | 10177 | -0.323 | -0.2089 | Yes |
| 98 | Hccs | 10422 | -0.349 | -0.2171 | Yes |
| 99 | Crat | 10479 | -0.354 | -0.2100 | Yes |
| 100 | Bckdhb | 10535 | -0.362 | -0.2025 | Yes |
| 101 | Suclg2 | 10624 | -0.371 | -0.1974 | Yes |
| 102 | Hibch | 10698 | -0.380 | -0.1908 | Yes |
| 103 | Acads | 10892 | -0.402 | -0.1932 | Yes |
| 104 | Sdhd | 11104 | -0.432 | -0.1961 | Yes |
| 105 | Acsl1 | 11284 | -0.457 | -0.1955 | Yes |
| 106 | Acadl | 11307 | -0.460 | -0.1821 | Yes |
| 107 | Ehhadh | 11391 | -0.474 | -0.1732 | Yes |
| 108 | Idh1 | 11793 | -0.557 | -0.1874 | Yes |
| 109 | Mdh2 | 12043 | -0.633 | -0.1868 | Yes |
| 110 | Hsd17b7 | 12055 | -0.635 | -0.1667 | Yes |
| 111 | Hsd17b11 | 12083 | -0.647 | -0.1475 | Yes |
| 112 | Hsd17b10 | 12150 | -0.679 | -0.1305 | Yes |
| 113 | Eci2 | 12176 | -0.696 | -0.1096 | Yes |
| 114 | Hsdl2 | 12229 | -0.722 | -0.0900 | Yes |
| 115 | Ostc | 12250 | -0.744 | -0.0670 | Yes |
| 116 | Ldha | 12252 | -0.745 | -0.0425 | Yes |
| 117 | Aldh9a1 | 12274 | -0.767 | -0.0189 | Yes |
| 118 | Acaa2 | 12402 | -0.982 | 0.0032 | Yes |