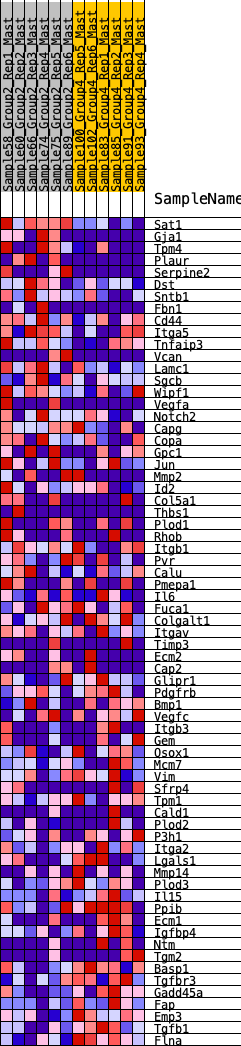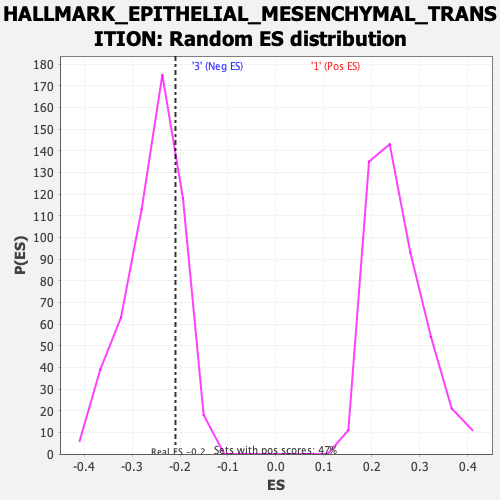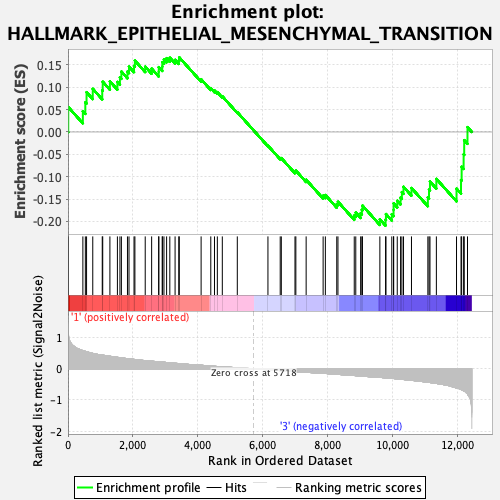
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.20950368 |
| Normalized Enrichment Score (NES) | -0.8213254 |
| Nominal p-value | 0.768797 |
| FDR q-value | 0.9221063 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sat1 | 8 | 1.214 | 0.0556 | No |
| 2 | Gja1 | 458 | 0.585 | 0.0465 | No |
| 3 | Tpm4 | 534 | 0.561 | 0.0664 | No |
| 4 | Plaur | 572 | 0.550 | 0.0889 | No |
| 5 | Serpine2 | 763 | 0.496 | 0.0966 | No |
| 6 | Dst | 1055 | 0.438 | 0.0933 | No |
| 7 | Sntb1 | 1070 | 0.436 | 0.1124 | No |
| 8 | Fbn1 | 1291 | 0.404 | 0.1134 | No |
| 9 | Cd44 | 1522 | 0.371 | 0.1120 | No |
| 10 | Itga5 | 1600 | 0.360 | 0.1224 | No |
| 11 | Tnfaip3 | 1645 | 0.352 | 0.1352 | No |
| 12 | Vcan | 1832 | 0.325 | 0.1352 | No |
| 13 | Lamc1 | 1879 | 0.319 | 0.1463 | No |
| 14 | Sgcb | 2034 | 0.303 | 0.1479 | No |
| 15 | Wipf1 | 2061 | 0.299 | 0.1597 | No |
| 16 | Vegfa | 2379 | 0.264 | 0.1463 | No |
| 17 | Notch2 | 2577 | 0.245 | 0.1417 | No |
| 18 | Capg | 2795 | 0.226 | 0.1347 | No |
| 19 | Copa | 2797 | 0.226 | 0.1451 | No |
| 20 | Gpc1 | 2904 | 0.216 | 0.1465 | No |
| 21 | Jun | 2908 | 0.216 | 0.1562 | No |
| 22 | Mmp2 | 2956 | 0.211 | 0.1622 | No |
| 23 | Id2 | 3038 | 0.203 | 0.1651 | No |
| 24 | Col5a1 | 3137 | 0.194 | 0.1662 | No |
| 25 | Thbs1 | 3299 | 0.178 | 0.1614 | No |
| 26 | Plod1 | 3413 | 0.171 | 0.1602 | No |
| 27 | Rhob | 3428 | 0.169 | 0.1669 | No |
| 28 | Itgb1 | 4101 | 0.115 | 0.1179 | No |
| 29 | Pvr | 4399 | 0.090 | 0.0981 | No |
| 30 | Calu | 4513 | 0.081 | 0.0927 | No |
| 31 | Pmepa1 | 4601 | 0.074 | 0.0891 | No |
| 32 | Il6 | 4754 | 0.064 | 0.0798 | No |
| 33 | Fuca1 | 5216 | 0.035 | 0.0442 | No |
| 34 | Colgalt1 | 6160 | -0.030 | -0.0306 | No |
| 35 | Itgav | 6538 | -0.056 | -0.0585 | No |
| 36 | Timp3 | 6572 | -0.058 | -0.0585 | No |
| 37 | Ecm2 | 6993 | -0.086 | -0.0884 | No |
| 38 | Cap2 | 7020 | -0.088 | -0.0865 | No |
| 39 | Glipr1 | 7337 | -0.110 | -0.1069 | No |
| 40 | Pdgfrb | 7860 | -0.143 | -0.1425 | No |
| 41 | Bmp1 | 7929 | -0.147 | -0.1411 | No |
| 42 | Vegfc | 8280 | -0.173 | -0.1614 | No |
| 43 | Itgb3 | 8317 | -0.176 | -0.1561 | No |
| 44 | Gem | 8819 | -0.212 | -0.1868 | No |
| 45 | Qsox1 | 8866 | -0.217 | -0.1804 | No |
| 46 | Mcm7 | 9016 | -0.228 | -0.1819 | No |
| 47 | Vim | 9053 | -0.231 | -0.1741 | No |
| 48 | Sfrp4 | 9072 | -0.233 | -0.1648 | No |
| 49 | Tpm1 | 9608 | -0.273 | -0.1954 | No |
| 50 | Cald1 | 9784 | -0.287 | -0.1962 | Yes |
| 51 | Plod2 | 9796 | -0.288 | -0.1837 | Yes |
| 52 | P3h1 | 9976 | -0.304 | -0.1841 | Yes |
| 53 | Itga2 | 10030 | -0.309 | -0.1741 | Yes |
| 54 | Lgals1 | 10033 | -0.309 | -0.1599 | Yes |
| 55 | Mmp14 | 10144 | -0.320 | -0.1540 | Yes |
| 56 | Plod3 | 10246 | -0.331 | -0.1468 | Yes |
| 57 | Il15 | 10287 | -0.335 | -0.1345 | Yes |
| 58 | Ppib | 10337 | -0.341 | -0.1227 | Yes |
| 59 | Ecm1 | 10582 | -0.367 | -0.1254 | Yes |
| 60 | Igfbp4 | 11087 | -0.431 | -0.1462 | Yes |
| 61 | Ntm | 11123 | -0.435 | -0.1288 | Yes |
| 62 | Tgm2 | 11152 | -0.438 | -0.1108 | Yes |
| 63 | Basp1 | 11349 | -0.467 | -0.1050 | Yes |
| 64 | Tgfbr3 | 11970 | -0.610 | -0.1269 | Yes |
| 65 | Gadd45a | 12109 | -0.660 | -0.1074 | Yes |
| 66 | Fap | 12128 | -0.669 | -0.0779 | Yes |
| 67 | Emp3 | 12189 | -0.704 | -0.0501 | Yes |
| 68 | Tgfb1 | 12207 | -0.714 | -0.0184 | Yes |
| 69 | Flna | 12308 | -0.803 | 0.0107 | Yes |