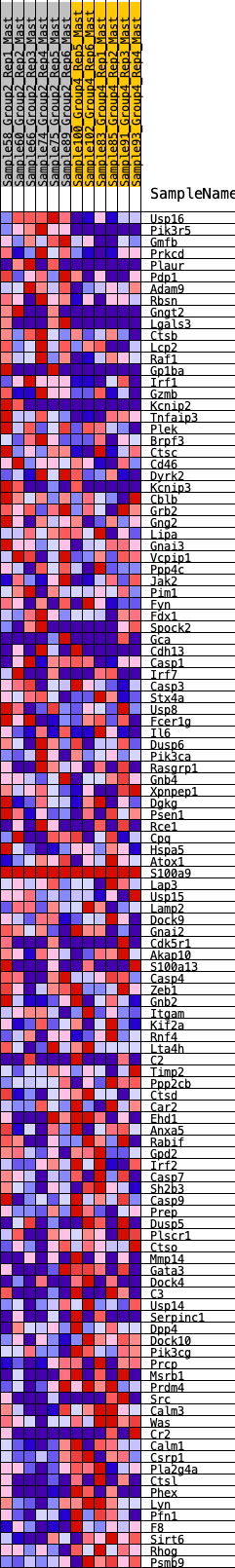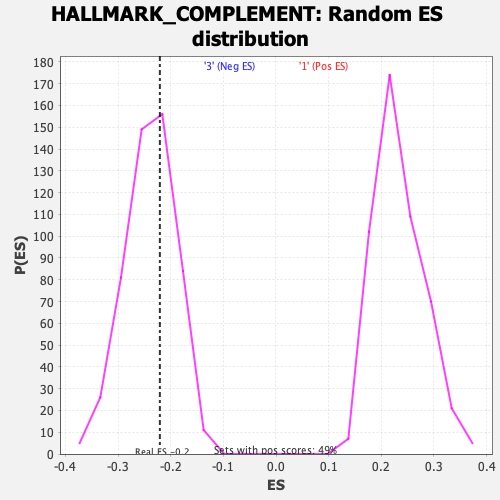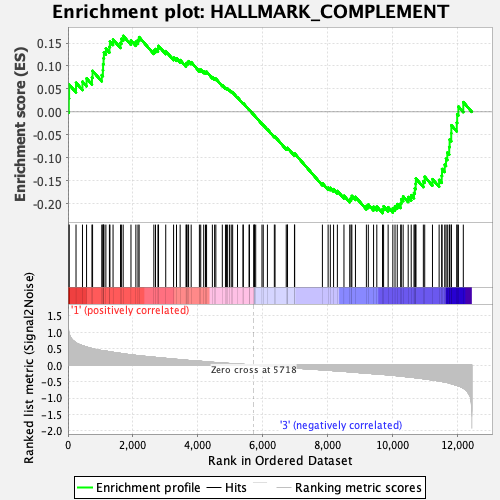
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.22068353 |
| Normalized Enrichment Score (NES) | -0.9253848 |
| Nominal p-value | 0.5996094 |
| FDR q-value | 0.92614967 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Usp16 | 25 | 1.016 | 0.0298 | No |
| 2 | Pik3r5 | 34 | 0.973 | 0.0596 | No |
| 3 | Gmfb | 247 | 0.678 | 0.0636 | No |
| 4 | Prkcd | 446 | 0.587 | 0.0659 | No |
| 5 | Plaur | 572 | 0.550 | 0.0729 | No |
| 6 | Pdp1 | 738 | 0.501 | 0.0752 | No |
| 7 | Adam9 | 757 | 0.497 | 0.0893 | No |
| 8 | Rbsn | 1041 | 0.441 | 0.0802 | No |
| 9 | Gngt2 | 1075 | 0.435 | 0.0911 | No |
| 10 | Lgals3 | 1081 | 0.435 | 0.1043 | No |
| 11 | Ctsb | 1096 | 0.434 | 0.1167 | No |
| 12 | Lcp2 | 1107 | 0.432 | 0.1295 | No |
| 13 | Raf1 | 1167 | 0.425 | 0.1380 | No |
| 14 | Gp1ba | 1276 | 0.406 | 0.1419 | No |
| 15 | Irf1 | 1292 | 0.404 | 0.1533 | No |
| 16 | Gzmb | 1388 | 0.389 | 0.1578 | No |
| 17 | Kcnip2 | 1618 | 0.356 | 0.1503 | No |
| 18 | Tnfaip3 | 1645 | 0.352 | 0.1592 | No |
| 19 | Plek | 1700 | 0.345 | 0.1656 | No |
| 20 | Brpf3 | 1940 | 0.313 | 0.1561 | No |
| 21 | Ctsc | 2091 | 0.295 | 0.1531 | No |
| 22 | Cd46 | 2156 | 0.286 | 0.1569 | No |
| 23 | Dyrk2 | 2191 | 0.282 | 0.1629 | No |
| 24 | Kcnip3 | 2645 | 0.239 | 0.1337 | No |
| 25 | Cblb | 2696 | 0.235 | 0.1369 | No |
| 26 | Grb2 | 2772 | 0.228 | 0.1380 | No |
| 27 | Gng2 | 2789 | 0.227 | 0.1438 | No |
| 28 | Lipa | 3011 | 0.206 | 0.1323 | No |
| 29 | Gnai3 | 3253 | 0.182 | 0.1185 | No |
| 30 | Vcpip1 | 3343 | 0.175 | 0.1167 | No |
| 31 | Ppp4c | 3455 | 0.167 | 0.1129 | No |
| 32 | Jak2 | 3636 | 0.150 | 0.1030 | No |
| 33 | Pim1 | 3651 | 0.148 | 0.1065 | No |
| 34 | Fyn | 3693 | 0.144 | 0.1077 | No |
| 35 | Fdx1 | 3715 | 0.142 | 0.1104 | No |
| 36 | Spock2 | 3795 | 0.136 | 0.1083 | No |
| 37 | Gca | 4048 | 0.119 | 0.0916 | No |
| 38 | Cdh13 | 4089 | 0.116 | 0.0920 | No |
| 39 | Casp1 | 4181 | 0.109 | 0.0880 | No |
| 40 | Irf7 | 4236 | 0.104 | 0.0869 | No |
| 41 | Casp3 | 4269 | 0.101 | 0.0874 | No |
| 42 | Stx4a | 4448 | 0.086 | 0.0757 | No |
| 43 | Usp8 | 4517 | 0.081 | 0.0727 | No |
| 44 | Fcer1g | 4555 | 0.079 | 0.0722 | No |
| 45 | Il6 | 4754 | 0.064 | 0.0581 | No |
| 46 | Dusp6 | 4851 | 0.058 | 0.0521 | No |
| 47 | Pik3ca | 4882 | 0.057 | 0.0515 | No |
| 48 | Rasgrp1 | 4918 | 0.054 | 0.0503 | No |
| 49 | Gnb4 | 4978 | 0.050 | 0.0471 | No |
| 50 | Xpnpep1 | 5029 | 0.047 | 0.0445 | No |
| 51 | Dgkg | 5073 | 0.044 | 0.0424 | No |
| 52 | Psen1 | 5222 | 0.035 | 0.0315 | No |
| 53 | Rce1 | 5391 | 0.025 | 0.0187 | No |
| 54 | Cpq | 5401 | 0.024 | 0.0187 | No |
| 55 | Hspa5 | 5579 | 0.012 | 0.0047 | No |
| 56 | Atox1 | 5590 | 0.011 | 0.0042 | No |
| 57 | S100a9 | 5721 | 0.000 | -0.0063 | No |
| 58 | Lap3 | 5741 | -0.000 | -0.0079 | No |
| 59 | Usp15 | 5744 | -0.000 | -0.0080 | No |
| 60 | Lamp2 | 5783 | -0.003 | -0.0110 | No |
| 61 | Dock9 | 5976 | -0.016 | -0.0261 | No |
| 62 | Gnai2 | 6015 | -0.019 | -0.0286 | No |
| 63 | Cdk5r1 | 6144 | -0.029 | -0.0380 | No |
| 64 | Akap10 | 6357 | -0.044 | -0.0539 | No |
| 65 | S100a13 | 6379 | -0.046 | -0.0541 | No |
| 66 | Casp4 | 6723 | -0.069 | -0.0798 | No |
| 67 | Zeb1 | 6756 | -0.071 | -0.0802 | No |
| 68 | Gnb2 | 6763 | -0.071 | -0.0785 | No |
| 69 | Itgam | 6977 | -0.085 | -0.0931 | No |
| 70 | Kif2a | 6984 | -0.086 | -0.0909 | No |
| 71 | Rnf4 | 7838 | -0.141 | -0.1557 | No |
| 72 | Lta4h | 8014 | -0.153 | -0.1651 | No |
| 73 | C2 | 8082 | -0.159 | -0.1655 | No |
| 74 | Timp2 | 8182 | -0.166 | -0.1684 | No |
| 75 | Ppp2cb | 8299 | -0.175 | -0.1723 | No |
| 76 | Ctsd | 8502 | -0.189 | -0.1828 | No |
| 77 | Car2 | 8680 | -0.202 | -0.1908 | No |
| 78 | Ehd1 | 8720 | -0.205 | -0.1876 | No |
| 79 | Anxa5 | 8745 | -0.207 | -0.1831 | No |
| 80 | Rabif | 8856 | -0.216 | -0.1852 | No |
| 81 | Gpd2 | 9193 | -0.243 | -0.2049 | No |
| 82 | Irf2 | 9252 | -0.248 | -0.2018 | No |
| 83 | Casp7 | 9414 | -0.259 | -0.2068 | No |
| 84 | Sh2b3 | 9514 | -0.267 | -0.2065 | No |
| 85 | Casp9 | 9690 | -0.279 | -0.2119 | Yes |
| 86 | Prep | 9723 | -0.282 | -0.2057 | Yes |
| 87 | Dusp5 | 9863 | -0.294 | -0.2078 | Yes |
| 88 | Plscr1 | 10008 | -0.307 | -0.2099 | Yes |
| 89 | Ctso | 10075 | -0.313 | -0.2054 | Yes |
| 90 | Mmp14 | 10144 | -0.320 | -0.2009 | Yes |
| 91 | Gata3 | 10251 | -0.331 | -0.1992 | Yes |
| 92 | Dock4 | 10267 | -0.332 | -0.1900 | Yes |
| 93 | C3 | 10326 | -0.339 | -0.1841 | Yes |
| 94 | Usp14 | 10481 | -0.354 | -0.1855 | Yes |
| 95 | Serpinc1 | 10574 | -0.366 | -0.1815 | Yes |
| 96 | Dpp4 | 10660 | -0.375 | -0.1767 | Yes |
| 97 | Dock10 | 10685 | -0.379 | -0.1668 | Yes |
| 98 | Pik3cg | 10711 | -0.382 | -0.1568 | Yes |
| 99 | Prcp | 10718 | -0.384 | -0.1453 | Yes |
| 100 | Msrb1 | 10948 | -0.411 | -0.1510 | Yes |
| 101 | Prdm4 | 10990 | -0.416 | -0.1414 | Yes |
| 102 | Src | 11229 | -0.447 | -0.1467 | Yes |
| 103 | Calm3 | 11434 | -0.481 | -0.1482 | Yes |
| 104 | Was | 11514 | -0.493 | -0.1392 | Yes |
| 105 | Cr2 | 11524 | -0.495 | -0.1244 | Yes |
| 106 | Calm1 | 11607 | -0.512 | -0.1151 | Yes |
| 107 | Csrp1 | 11646 | -0.519 | -0.1019 | Yes |
| 108 | Pla2g4a | 11686 | -0.526 | -0.0886 | Yes |
| 109 | Ctsl | 11745 | -0.542 | -0.0764 | Yes |
| 110 | Phex | 11758 | -0.546 | -0.0602 | Yes |
| 111 | Lyn | 11807 | -0.562 | -0.0466 | Yes |
| 112 | Pfn1 | 11811 | -0.563 | -0.0292 | Yes |
| 113 | F8 | 11978 | -0.613 | -0.0235 | Yes |
| 114 | Sirt6 | 11991 | -0.618 | -0.0051 | Yes |
| 115 | Rhog | 12028 | -0.628 | 0.0116 | Yes |
| 116 | Psmb9 | 12179 | -0.698 | 0.0213 | Yes |