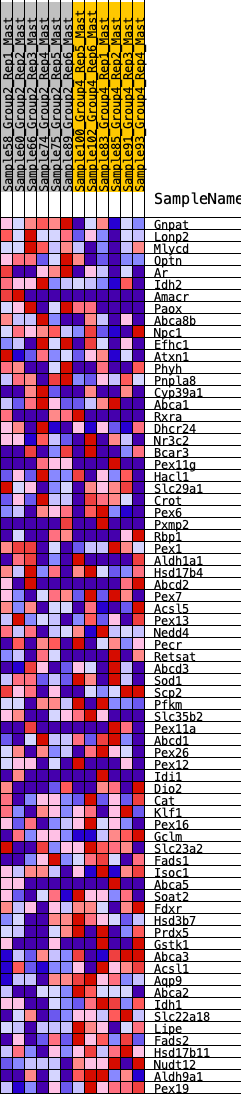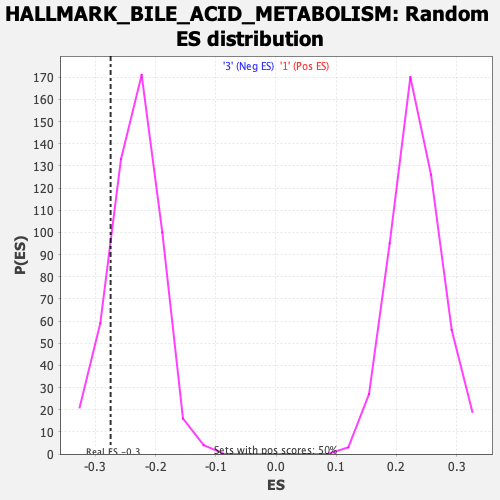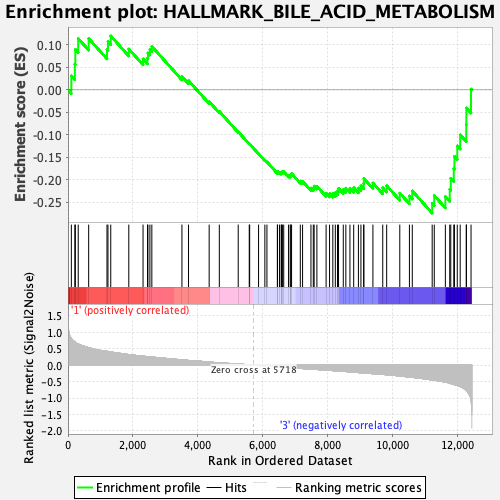
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.274666 |
| Normalized Enrichment Score (NES) | -1.1663748 |
| Nominal p-value | 0.15873016 |
| FDR q-value | 0.75489503 |
| FWER p-Value | 0.927 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gnpat | 103 | 0.807 | 0.0313 | No |
| 2 | Lonp2 | 211 | 0.706 | 0.0573 | No |
| 3 | Mlycd | 226 | 0.690 | 0.0901 | No |
| 4 | Optn | 315 | 0.640 | 0.1144 | No |
| 5 | Ar | 637 | 0.531 | 0.1145 | No |
| 6 | Idh2 | 1201 | 0.420 | 0.0896 | No |
| 7 | Amacr | 1231 | 0.412 | 0.1075 | No |
| 8 | Paox | 1317 | 0.400 | 0.1203 | No |
| 9 | Abca8b | 1873 | 0.320 | 0.0911 | No |
| 10 | Npc1 | 2312 | 0.270 | 0.0690 | No |
| 11 | Efhc1 | 2450 | 0.258 | 0.0706 | No |
| 12 | Atxn1 | 2462 | 0.256 | 0.0823 | No |
| 13 | Phyh | 2523 | 0.250 | 0.0897 | No |
| 14 | Pnpla8 | 2583 | 0.245 | 0.0969 | No |
| 15 | Cyp39a1 | 3510 | 0.161 | 0.0299 | No |
| 16 | Abca1 | 3713 | 0.142 | 0.0206 | No |
| 17 | Rxra | 4349 | 0.094 | -0.0261 | No |
| 18 | Dhcr24 | 4662 | 0.070 | -0.0479 | No |
| 19 | Nr3c2 | 5244 | 0.034 | -0.0933 | No |
| 20 | Bcar3 | 5584 | 0.011 | -0.1201 | No |
| 21 | Pex11g | 5597 | 0.010 | -0.1206 | No |
| 22 | Hacl1 | 5871 | -0.010 | -0.1422 | No |
| 23 | Slc29a1 | 6065 | -0.023 | -0.1567 | No |
| 24 | Crot | 6129 | -0.028 | -0.1604 | No |
| 25 | Pex6 | 6451 | -0.050 | -0.1839 | No |
| 26 | Pxmp2 | 6453 | -0.050 | -0.1815 | No |
| 27 | Rbp1 | 6515 | -0.054 | -0.1838 | No |
| 28 | Pex1 | 6567 | -0.058 | -0.1850 | No |
| 29 | Aldh1a1 | 6578 | -0.058 | -0.1830 | No |
| 30 | Hsd17b4 | 6606 | -0.060 | -0.1822 | No |
| 31 | Abcd2 | 6646 | -0.063 | -0.1823 | No |
| 32 | Pex7 | 6795 | -0.074 | -0.1906 | No |
| 33 | Acsl5 | 6846 | -0.077 | -0.1909 | No |
| 34 | Pex13 | 6865 | -0.078 | -0.1885 | No |
| 35 | Nedd4 | 6888 | -0.079 | -0.1864 | No |
| 36 | Pecr | 7157 | -0.098 | -0.2032 | No |
| 37 | Retsat | 7227 | -0.103 | -0.2038 | No |
| 38 | Abcd3 | 7485 | -0.118 | -0.2187 | No |
| 39 | Sod1 | 7557 | -0.122 | -0.2185 | No |
| 40 | Scp2 | 7583 | -0.125 | -0.2144 | No |
| 41 | Pfkm | 7668 | -0.131 | -0.2147 | No |
| 42 | Slc35b2 | 7952 | -0.148 | -0.2303 | No |
| 43 | Pex11a | 8058 | -0.157 | -0.2311 | No |
| 44 | Abcd1 | 8159 | -0.164 | -0.2311 | No |
| 45 | Pex26 | 8236 | -0.170 | -0.2289 | No |
| 46 | Pex12 | 8303 | -0.175 | -0.2257 | No |
| 47 | Idi1 | 8335 | -0.178 | -0.2195 | No |
| 48 | Dio2 | 8481 | -0.187 | -0.2220 | No |
| 49 | Cat | 8560 | -0.192 | -0.2189 | No |
| 50 | Klf1 | 8686 | -0.203 | -0.2190 | No |
| 51 | Pex16 | 8802 | -0.211 | -0.2180 | No |
| 52 | Gclm | 8947 | -0.223 | -0.2186 | No |
| 53 | Slc23a2 | 9025 | -0.229 | -0.2136 | No |
| 54 | Fads1 | 9109 | -0.236 | -0.2088 | No |
| 55 | Isoc1 | 9116 | -0.237 | -0.1976 | No |
| 56 | Abca5 | 9394 | -0.258 | -0.2074 | No |
| 57 | Soat2 | 9697 | -0.280 | -0.2180 | No |
| 58 | Fdxr | 9818 | -0.290 | -0.2135 | No |
| 59 | Hsd3b7 | 10222 | -0.328 | -0.2300 | No |
| 60 | Prdx5 | 10519 | -0.360 | -0.2363 | No |
| 61 | Gstk1 | 10607 | -0.369 | -0.2252 | No |
| 62 | Abca3 | 11220 | -0.446 | -0.2528 | Yes |
| 63 | Acsl1 | 11284 | -0.457 | -0.2355 | Yes |
| 64 | Aqp9 | 11626 | -0.515 | -0.2377 | Yes |
| 65 | Abca2 | 11761 | -0.547 | -0.2217 | Yes |
| 66 | Idh1 | 11793 | -0.557 | -0.1969 | Yes |
| 67 | Slc22a18 | 11886 | -0.584 | -0.1757 | Yes |
| 68 | Lipe | 11906 | -0.592 | -0.1481 | Yes |
| 69 | Fads2 | 11993 | -0.618 | -0.1248 | Yes |
| 70 | Hsd17b11 | 12083 | -0.647 | -0.1002 | Yes |
| 71 | Nudt12 | 12271 | -0.765 | -0.0778 | Yes |
| 72 | Aldh9a1 | 12274 | -0.767 | -0.0403 | Yes |
| 73 | Pex19 | 12417 | -1.094 | 0.0019 | Yes |