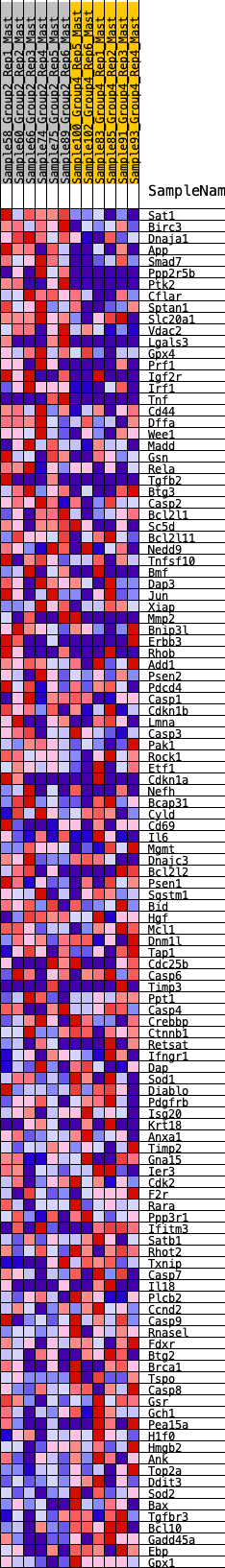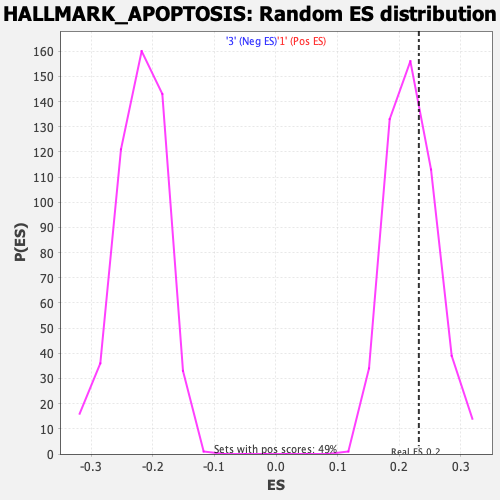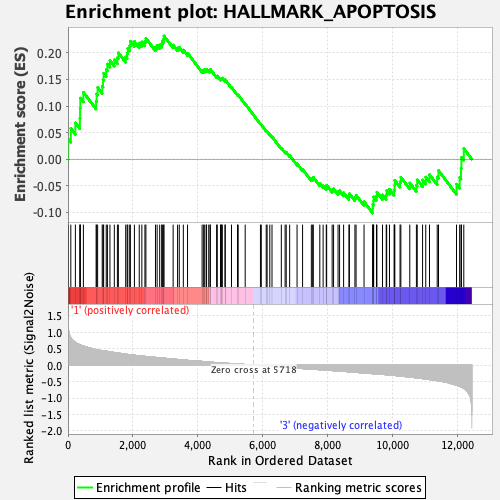
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.2319001 |
| Normalized Enrichment Score (NES) | 1.0551105 |
| Nominal p-value | 0.3510204 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sat1 | 8 | 1.214 | 0.0378 | Yes |
| 2 | Birc3 | 87 | 0.835 | 0.0580 | Yes |
| 3 | Dnaja1 | 227 | 0.688 | 0.0685 | Yes |
| 4 | App | 366 | 0.620 | 0.0770 | Yes |
| 5 | Smad7 | 373 | 0.618 | 0.0961 | Yes |
| 6 | Ppp2r5b | 380 | 0.614 | 0.1150 | Yes |
| 7 | Ptk2 | 474 | 0.580 | 0.1259 | Yes |
| 8 | Cflar | 866 | 0.473 | 0.1092 | Yes |
| 9 | Sptan1 | 883 | 0.469 | 0.1227 | Yes |
| 10 | Slc20a1 | 913 | 0.463 | 0.1350 | Yes |
| 11 | Vdac2 | 1054 | 0.438 | 0.1375 | Yes |
| 12 | Lgals3 | 1081 | 0.435 | 0.1492 | Yes |
| 13 | Gpx4 | 1098 | 0.434 | 0.1617 | Yes |
| 14 | Prf1 | 1178 | 0.424 | 0.1687 | Yes |
| 15 | Igf2r | 1214 | 0.418 | 0.1791 | Yes |
| 16 | Irf1 | 1292 | 0.404 | 0.1856 | Yes |
| 17 | Tnf | 1427 | 0.383 | 0.1869 | Yes |
| 18 | Cd44 | 1522 | 0.371 | 0.1911 | Yes |
| 19 | Dffa | 1552 | 0.366 | 0.2003 | Yes |
| 20 | Wee1 | 1778 | 0.333 | 0.1926 | Yes |
| 21 | Madd | 1822 | 0.326 | 0.1995 | Yes |
| 22 | Gsn | 1842 | 0.325 | 0.2082 | Yes |
| 23 | Rela | 1902 | 0.317 | 0.2135 | Yes |
| 24 | Tgfb2 | 1921 | 0.314 | 0.2220 | Yes |
| 25 | Btg3 | 2046 | 0.302 | 0.2215 | Yes |
| 26 | Casp2 | 2195 | 0.282 | 0.2184 | Yes |
| 27 | Bcl2l1 | 2275 | 0.274 | 0.2207 | Yes |
| 28 | Sc5d | 2370 | 0.265 | 0.2215 | Yes |
| 29 | Bcl2l11 | 2400 | 0.263 | 0.2274 | Yes |
| 30 | Nedd9 | 2695 | 0.235 | 0.2110 | Yes |
| 31 | Tnfsf10 | 2746 | 0.230 | 0.2143 | Yes |
| 32 | Bmf | 2825 | 0.224 | 0.2150 | Yes |
| 33 | Dap3 | 2886 | 0.218 | 0.2171 | Yes |
| 34 | Jun | 2908 | 0.216 | 0.2222 | Yes |
| 35 | Xiap | 2948 | 0.212 | 0.2258 | Yes |
| 36 | Mmp2 | 2956 | 0.211 | 0.2319 | Yes |
| 37 | Bnip3l | 3239 | 0.184 | 0.2148 | No |
| 38 | Erbb3 | 3376 | 0.173 | 0.2093 | No |
| 39 | Rhob | 3428 | 0.169 | 0.2105 | No |
| 40 | Add1 | 3553 | 0.158 | 0.2055 | No |
| 41 | Psen2 | 3683 | 0.145 | 0.1996 | No |
| 42 | Pdcd4 | 4132 | 0.112 | 0.1668 | No |
| 43 | Casp1 | 4181 | 0.109 | 0.1664 | No |
| 44 | Cdkn1b | 4198 | 0.107 | 0.1685 | No |
| 45 | Lmna | 4255 | 0.102 | 0.1671 | No |
| 46 | Casp3 | 4269 | 0.101 | 0.1693 | No |
| 47 | Pak1 | 4336 | 0.095 | 0.1669 | No |
| 48 | Rock1 | 4378 | 0.092 | 0.1665 | No |
| 49 | Etf1 | 4384 | 0.091 | 0.1690 | No |
| 50 | Cdkn1a | 4579 | 0.077 | 0.1557 | No |
| 51 | Nefh | 4598 | 0.075 | 0.1566 | No |
| 52 | Bcap31 | 4699 | 0.068 | 0.1506 | No |
| 53 | Cyld | 4718 | 0.067 | 0.1513 | No |
| 54 | Cd69 | 4734 | 0.065 | 0.1521 | No |
| 55 | Il6 | 4754 | 0.064 | 0.1526 | No |
| 56 | Mgmt | 4839 | 0.059 | 0.1477 | No |
| 57 | Dnajc3 | 4843 | 0.059 | 0.1493 | No |
| 58 | Bcl2l2 | 5037 | 0.047 | 0.1351 | No |
| 59 | Psen1 | 5222 | 0.035 | 0.1213 | No |
| 60 | Sqstm1 | 5243 | 0.034 | 0.1208 | No |
| 61 | Bid | 5459 | 0.020 | 0.1039 | No |
| 62 | Hgf | 5929 | -0.014 | 0.0663 | No |
| 63 | Mcl1 | 5955 | -0.015 | 0.0648 | No |
| 64 | Dnm1l | 6107 | -0.027 | 0.0534 | No |
| 65 | Tap1 | 6134 | -0.028 | 0.0522 | No |
| 66 | Cdc25b | 6217 | -0.034 | 0.0466 | No |
| 67 | Casp6 | 6285 | -0.039 | 0.0424 | No |
| 68 | Timp3 | 6572 | -0.058 | 0.0210 | No |
| 69 | Ppt1 | 6689 | -0.066 | 0.0137 | No |
| 70 | Casp4 | 6723 | -0.069 | 0.0132 | No |
| 71 | Crebbp | 6829 | -0.076 | 0.0071 | No |
| 72 | Ctnnb1 | 7057 | -0.091 | -0.0084 | No |
| 73 | Retsat | 7227 | -0.103 | -0.0189 | No |
| 74 | Ifngr1 | 7497 | -0.119 | -0.0369 | No |
| 75 | Dap | 7538 | -0.121 | -0.0363 | No |
| 76 | Sod1 | 7557 | -0.122 | -0.0339 | No |
| 77 | Diablo | 7758 | -0.136 | -0.0458 | No |
| 78 | Pdgfrb | 7860 | -0.143 | -0.0495 | No |
| 79 | Isg20 | 7957 | -0.148 | -0.0526 | No |
| 80 | Krt18 | 7969 | -0.149 | -0.0488 | No |
| 81 | Anxa1 | 8138 | -0.163 | -0.0572 | No |
| 82 | Timp2 | 8182 | -0.166 | -0.0555 | No |
| 83 | Gna15 | 8320 | -0.177 | -0.0610 | No |
| 84 | Ier3 | 8367 | -0.180 | -0.0590 | No |
| 85 | Cdk2 | 8491 | -0.188 | -0.0630 | No |
| 86 | F2r | 8656 | -0.200 | -0.0700 | No |
| 87 | Rara | 8669 | -0.201 | -0.0646 | No |
| 88 | Ppp3r1 | 8840 | -0.214 | -0.0716 | No |
| 89 | Ifitm3 | 8875 | -0.218 | -0.0675 | No |
| 90 | Satb1 | 9121 | -0.238 | -0.0798 | No |
| 91 | Rhot2 | 9388 | -0.257 | -0.0932 | No |
| 92 | Txnip | 9389 | -0.257 | -0.0851 | No |
| 93 | Casp7 | 9414 | -0.259 | -0.0788 | No |
| 94 | Il18 | 9416 | -0.259 | -0.0707 | No |
| 95 | Plcb2 | 9504 | -0.266 | -0.0693 | No |
| 96 | Ccnd2 | 9517 | -0.267 | -0.0618 | No |
| 97 | Casp9 | 9690 | -0.279 | -0.0669 | No |
| 98 | Rnasel | 9804 | -0.289 | -0.0669 | No |
| 99 | Fdxr | 9818 | -0.290 | -0.0588 | No |
| 100 | Btg2 | 9904 | -0.298 | -0.0563 | No |
| 101 | Brca1 | 10047 | -0.311 | -0.0579 | No |
| 102 | Tspo | 10063 | -0.312 | -0.0493 | No |
| 103 | Casp8 | 10066 | -0.312 | -0.0395 | No |
| 104 | Gsr | 10230 | -0.329 | -0.0423 | No |
| 105 | Gch1 | 10255 | -0.331 | -0.0338 | No |
| 106 | Pea15a | 10530 | -0.361 | -0.0446 | No |
| 107 | H1f0 | 10734 | -0.385 | -0.0489 | No |
| 108 | Hmgb2 | 10759 | -0.388 | -0.0385 | No |
| 109 | Ank | 10924 | -0.407 | -0.0389 | No |
| 110 | Top2a | 11022 | -0.421 | -0.0334 | No |
| 111 | Ddit3 | 11137 | -0.437 | -0.0289 | No |
| 112 | Sod2 | 11375 | -0.471 | -0.0332 | No |
| 113 | Bax | 11415 | -0.477 | -0.0212 | No |
| 114 | Tgfbr3 | 11970 | -0.610 | -0.0468 | No |
| 115 | Bcl10 | 12067 | -0.641 | -0.0343 | No |
| 116 | Gadd45a | 12109 | -0.660 | -0.0167 | No |
| 117 | Ebp | 12121 | -0.667 | 0.0035 | No |
| 118 | Gpx1 | 12194 | -0.706 | 0.0200 | No |