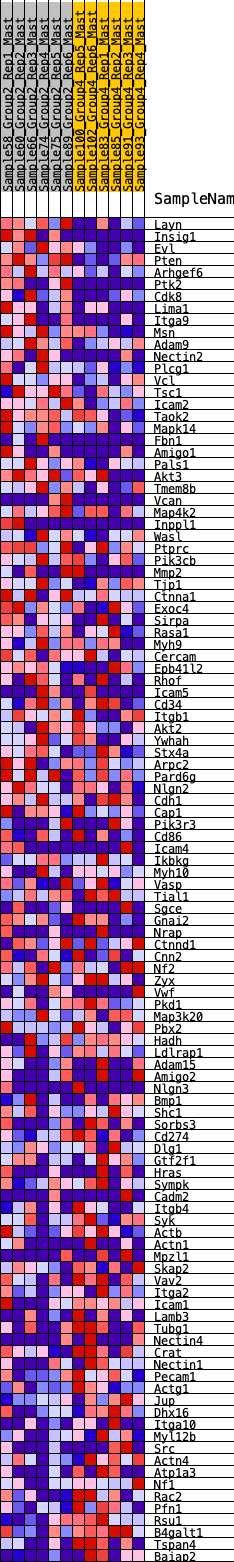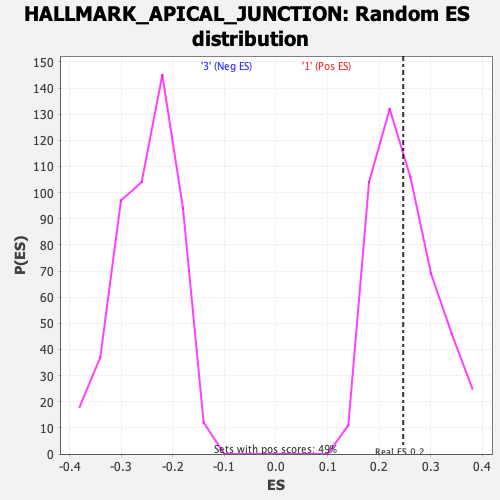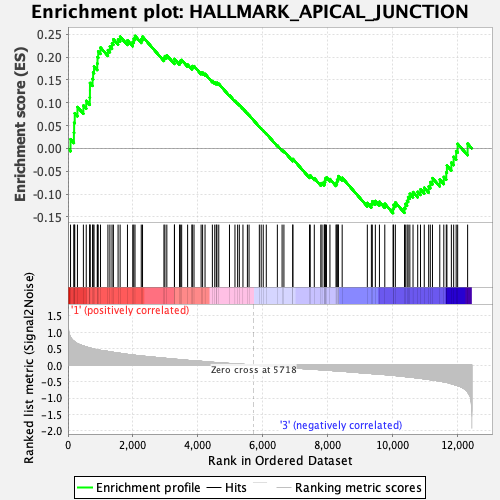
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Mast.Mast_Pheno.cls #Group2_versus_Group4.Mast_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Mast_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.24648206 |
| Normalized Enrichment Score (NES) | 0.98876774 |
| Nominal p-value | 0.46653143 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Layn | 78 | 0.847 | 0.0201 | Yes |
| 2 | Insig1 | 182 | 0.727 | 0.0344 | Yes |
| 3 | Evl | 189 | 0.721 | 0.0564 | Yes |
| 4 | Pten | 208 | 0.709 | 0.0770 | Yes |
| 5 | Arhgef6 | 291 | 0.651 | 0.0906 | Yes |
| 6 | Ptk2 | 474 | 0.580 | 0.0940 | Yes |
| 7 | Cdk8 | 562 | 0.553 | 0.1042 | Yes |
| 8 | Lima1 | 671 | 0.517 | 0.1115 | Yes |
| 9 | Itga9 | 676 | 0.516 | 0.1273 | Yes |
| 10 | Msn | 677 | 0.516 | 0.1434 | Yes |
| 11 | Adam9 | 757 | 0.497 | 0.1525 | Yes |
| 12 | Nectin2 | 774 | 0.494 | 0.1666 | Yes |
| 13 | Plcg1 | 802 | 0.486 | 0.1795 | Yes |
| 14 | Vcl | 901 | 0.465 | 0.1861 | Yes |
| 15 | Tsc1 | 909 | 0.464 | 0.2000 | Yes |
| 16 | Icam2 | 931 | 0.460 | 0.2126 | Yes |
| 17 | Taok2 | 1001 | 0.447 | 0.2209 | Yes |
| 18 | Mapk14 | 1230 | 0.412 | 0.2153 | Yes |
| 19 | Fbn1 | 1291 | 0.404 | 0.2230 | Yes |
| 20 | Amigo1 | 1356 | 0.395 | 0.2302 | Yes |
| 21 | Pals1 | 1398 | 0.388 | 0.2389 | Yes |
| 22 | Akt3 | 1543 | 0.367 | 0.2387 | Yes |
| 23 | Tmem8b | 1606 | 0.358 | 0.2448 | Yes |
| 24 | Vcan | 1832 | 0.325 | 0.2367 | Yes |
| 25 | Map4k2 | 1995 | 0.308 | 0.2332 | Yes |
| 26 | Inppl1 | 2026 | 0.304 | 0.2402 | Yes |
| 27 | Wasl | 2064 | 0.298 | 0.2465 | Yes |
| 28 | Ptprc | 2257 | 0.276 | 0.2395 | No |
| 29 | Pik3cb | 2296 | 0.272 | 0.2449 | No |
| 30 | Mmp2 | 2956 | 0.211 | 0.1980 | No |
| 31 | Tjp1 | 2986 | 0.209 | 0.2022 | No |
| 32 | Ctnna1 | 3045 | 0.201 | 0.2038 | No |
| 33 | Exoc4 | 3278 | 0.180 | 0.1905 | No |
| 34 | Sirpa | 3281 | 0.179 | 0.1960 | No |
| 35 | Rasa1 | 3437 | 0.168 | 0.1886 | No |
| 36 | Myh9 | 3460 | 0.166 | 0.1920 | No |
| 37 | Cercam | 3502 | 0.161 | 0.1937 | No |
| 38 | Epb41l2 | 3685 | 0.145 | 0.1835 | No |
| 39 | Rhof | 3819 | 0.135 | 0.1769 | No |
| 40 | Icam5 | 3828 | 0.134 | 0.1804 | No |
| 41 | Cd34 | 3882 | 0.130 | 0.1802 | No |
| 42 | Itgb1 | 4101 | 0.115 | 0.1661 | No |
| 43 | Akt2 | 4147 | 0.111 | 0.1659 | No |
| 44 | Ywhah | 4222 | 0.105 | 0.1632 | No |
| 45 | Stx4a | 4448 | 0.086 | 0.1476 | No |
| 46 | Arpc2 | 4521 | 0.081 | 0.1443 | No |
| 47 | Pard6g | 4569 | 0.078 | 0.1429 | No |
| 48 | Nlgn2 | 4588 | 0.076 | 0.1438 | No |
| 49 | Cdh1 | 4645 | 0.071 | 0.1415 | No |
| 50 | Cap1 | 4975 | 0.050 | 0.1164 | No |
| 51 | Pik3r3 | 5144 | 0.040 | 0.1040 | No |
| 52 | Cd86 | 5226 | 0.035 | 0.0985 | No |
| 53 | Icam4 | 5278 | 0.031 | 0.0953 | No |
| 54 | Ikbkg | 5390 | 0.025 | 0.0871 | No |
| 55 | Myh10 | 5527 | 0.015 | 0.0765 | No |
| 56 | Vasp | 5578 | 0.012 | 0.0728 | No |
| 57 | Tial1 | 5895 | -0.011 | 0.0476 | No |
| 58 | Sgce | 5952 | -0.015 | 0.0435 | No |
| 59 | Gnai2 | 6015 | -0.019 | 0.0391 | No |
| 60 | Nrap | 6109 | -0.027 | 0.0324 | No |
| 61 | Ctnnd1 | 6450 | -0.050 | 0.0063 | No |
| 62 | Cnn2 | 6596 | -0.059 | -0.0036 | No |
| 63 | Nf2 | 6652 | -0.063 | -0.0061 | No |
| 64 | Zyx | 6925 | -0.082 | -0.0256 | No |
| 65 | Vwf | 6927 | -0.082 | -0.0231 | No |
| 66 | Pkd1 | 7441 | -0.116 | -0.0611 | No |
| 67 | Map3k20 | 7463 | -0.117 | -0.0591 | No |
| 68 | Pbx2 | 7588 | -0.125 | -0.0653 | No |
| 69 | Hadh | 7789 | -0.138 | -0.0772 | No |
| 70 | Ldlrap1 | 7826 | -0.141 | -0.0758 | No |
| 71 | Adam15 | 7893 | -0.145 | -0.0766 | No |
| 72 | Amigo2 | 7907 | -0.146 | -0.0731 | No |
| 73 | Nlgn3 | 7921 | -0.147 | -0.0696 | No |
| 74 | Bmp1 | 7929 | -0.147 | -0.0655 | No |
| 75 | Shc1 | 7963 | -0.149 | -0.0636 | No |
| 76 | Sorbs3 | 8070 | -0.158 | -0.0672 | No |
| 77 | Cd274 | 8257 | -0.171 | -0.0770 | No |
| 78 | Dlg1 | 8284 | -0.174 | -0.0737 | No |
| 79 | Gtf2f1 | 8292 | -0.174 | -0.0688 | No |
| 80 | Hras | 8321 | -0.177 | -0.0656 | No |
| 81 | Sympk | 8330 | -0.177 | -0.0607 | No |
| 82 | Cadm2 | 8447 | -0.185 | -0.0643 | No |
| 83 | Itgb4 | 9222 | -0.246 | -0.1195 | No |
| 84 | Syk | 9347 | -0.255 | -0.1216 | No |
| 85 | Actb | 9375 | -0.256 | -0.1158 | No |
| 86 | Actn1 | 9469 | -0.263 | -0.1151 | No |
| 87 | Mpzl1 | 9595 | -0.272 | -0.1168 | No |
| 88 | Skap2 | 9760 | -0.285 | -0.1212 | No |
| 89 | Vav2 | 10015 | -0.307 | -0.1322 | No |
| 90 | Itga2 | 10030 | -0.309 | -0.1237 | No |
| 91 | Icam1 | 10085 | -0.314 | -0.1183 | No |
| 92 | Lamb3 | 10367 | -0.344 | -0.1304 | No |
| 93 | Tubg1 | 10392 | -0.347 | -0.1215 | No |
| 94 | Nectin4 | 10446 | -0.352 | -0.1149 | No |
| 95 | Crat | 10479 | -0.354 | -0.1064 | No |
| 96 | Nectin1 | 10532 | -0.361 | -0.0994 | No |
| 97 | Pecam1 | 10630 | -0.372 | -0.0956 | No |
| 98 | Actg1 | 10774 | -0.389 | -0.0951 | No |
| 99 | Jup | 10860 | -0.399 | -0.0896 | No |
| 100 | Dhx16 | 10976 | -0.415 | -0.0860 | No |
| 101 | Itga10 | 11112 | -0.434 | -0.0834 | No |
| 102 | Myl12b | 11162 | -0.439 | -0.0737 | No |
| 103 | Src | 11229 | -0.447 | -0.0651 | No |
| 104 | Actn4 | 11454 | -0.484 | -0.0682 | No |
| 105 | Atp1a3 | 11578 | -0.506 | -0.0624 | No |
| 106 | Nf1 | 11654 | -0.521 | -0.0523 | No |
| 107 | Rac2 | 11674 | -0.524 | -0.0375 | No |
| 108 | Pfn1 | 11811 | -0.563 | -0.0309 | No |
| 109 | Rsu1 | 11882 | -0.582 | -0.0185 | No |
| 110 | B4galt1 | 11959 | -0.607 | -0.0058 | No |
| 111 | Tspan4 | 12005 | -0.622 | 0.0100 | No |
| 112 | Baiap2 | 12312 | -0.812 | 0.0105 | No |